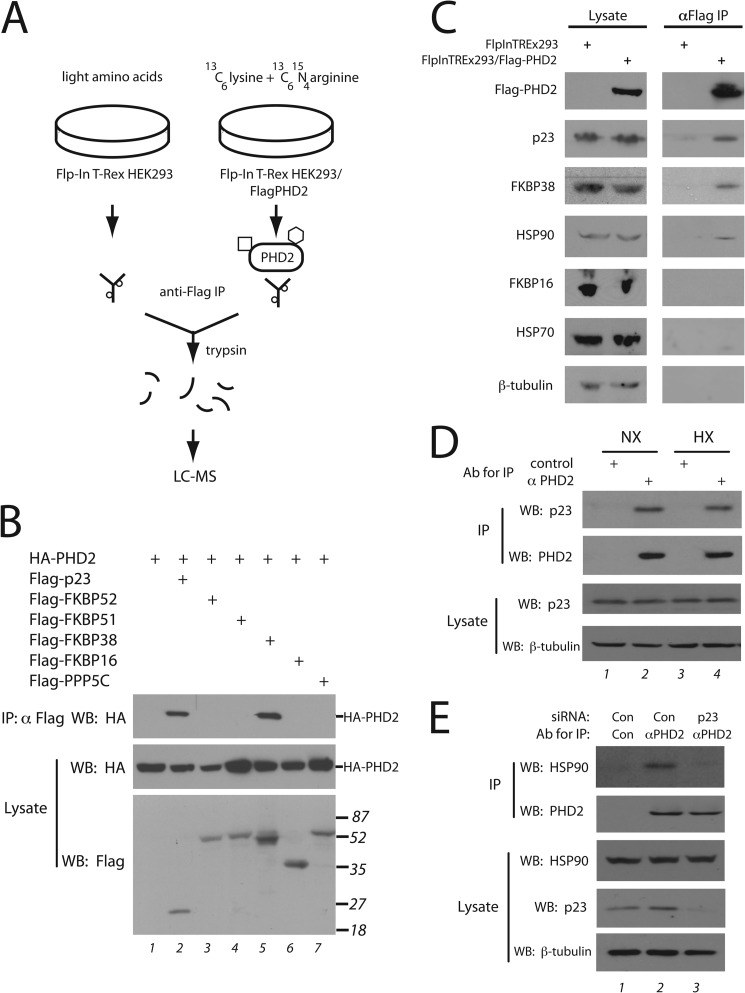
FIGURE 1.
Mass spectrometry identification of PHD2-interacting proteins. A, Flp-In T-Rex HEK293 or Flp-In T-Rex HEK293/FLAG-PHD2 cells were grown in the presence of amino acids with light or heavy labeled lysine/arginine, respectively and subjected to immunoprecipitation (IP) with anti-FLAG antibodies, and the eluates were mixed and subjected to tryptic digest and mass spectrometry. The square and hexagon depict proteins that bind specifically to PHD2, whereas the small circles depict proteins that bind non-specifically to the antibody (or resin). B, HEK293FT cells were transfected with expression vectors for the indicated proteins, lysed, and subjected to immunoprecipitation with anti-FLAG antibodies, and Western blots (WB) were then performed. The positions of HA-PHD2 or molecular weight markers are shown to the right. C, Flp-In T-Rex HEK293 or Flp-In T-Rex HEK293/FLAG-PHD2 cells were induced with doxycycline, lysed, and subjected to immunoprecipitation with anti-FLAG antibodies. The immunoprecipitates were eluted with 3X FLAG peptide. Aliquots of lysate or eluate were then subjected to Western blotting using antibodies against the indicated proteins. Anti-FLAG antibodies were employed to detect FLAG-PHD2. D and E, HEK293FT cells were maintained under normoxia or hypoxia (1% O2 for 4 h) (D) or treated with control or p23 siRNA (E). Ab, antibody. Cells were lysed, incubated with 10 μg of either control or anti-PHD2 monoclonal antibody, and immunoprecipitated with protein G-agarose, and Western blots were then performed.