Fig. 1.
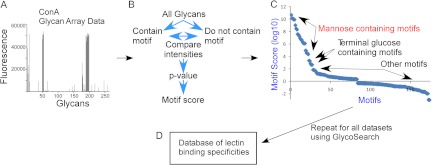
The development of the database of lectin-motif interactions. Quantified glycan array data (A) are processed using the GlycoSearch software and the motif segregation algorithm to produce motif scores for each experiment (B). The analysis of a glycan array experiment of the lectin concanavalin A (ConA) is given as an example, in which the top-scoring motifs contain mannose, followed by motifs with terminal glucose (C). All glycan array datasets were downloaded from the CFG website, processed, and assembled into a database (D). A variety of query types can be applied to the database.
