Figure 2.
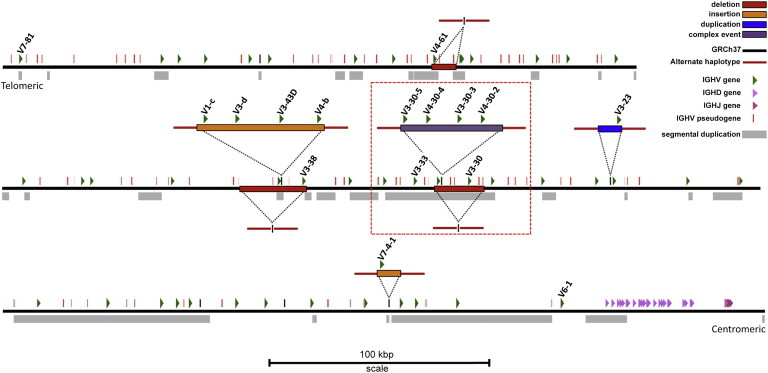
Map of IGHV CNVs Identified by Complete Sequencing of Fosmid Clones
The positions of three deletions, two insertions, one duplication, and one complex event characterized from fosmid alternative haplotypes are shown mapped to the GRCh37 IGH reference (black line; chr14:105,928,955–107,289,540) with the same parameters as in Figure 1B. The locus is presented in the same orientation as that depicted by IMGT. Functional and ORF IGHV, IGHD, and IGHJ genes (not to scale) as well as IGHV pseudogenes are shown (to scale); the names of IGHV genes involved in the characterized structural variants are indicated. Segmental duplications82,83 downloaded from the UCSC genome browser are shown below GRCh37. The large red box indicates a hotspot region of recurrent mutation (see Figure 4 for additional haplotypes associated with this hotspot). The deletion of IGHV4-61 was identified by Mills et al.5 (chr14:107,084,861–107,096,738) and was also included in our analyses.
