Figure S1.
Sequence Alignment of the Plasmodium spp SHLPs, Related to Figure 1
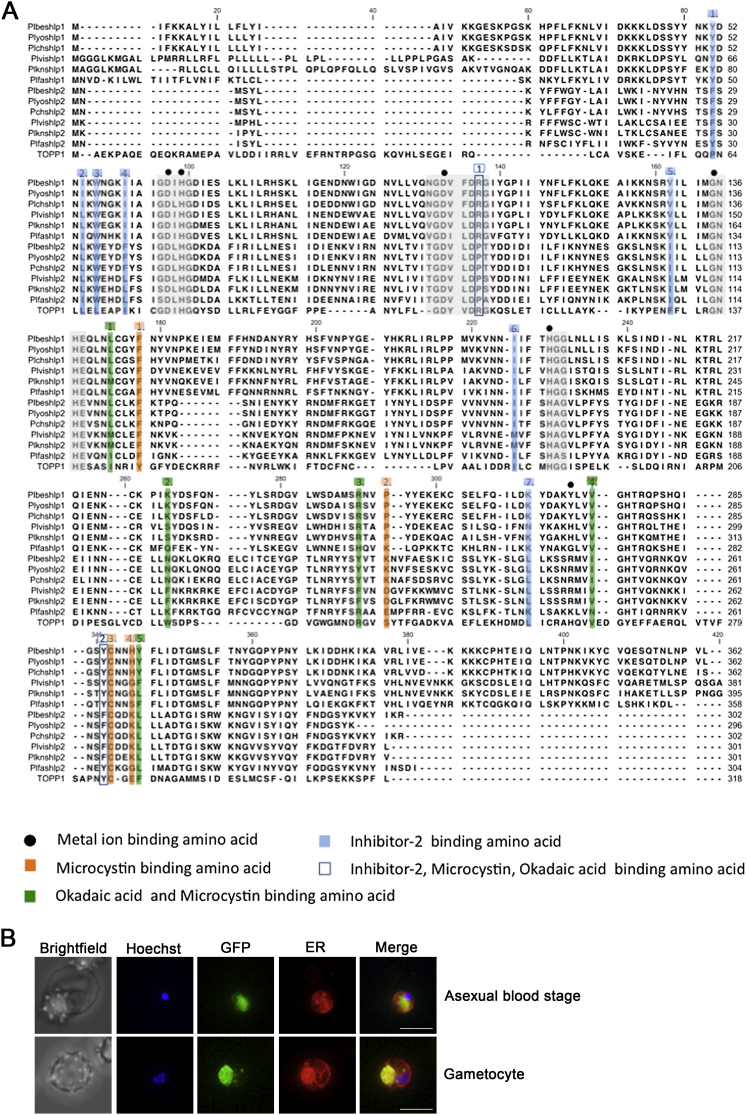
(A) The invariant residues conserved in protein phosphatases are highlighted in gray and metal ion binding sites are marked with solid circles. The amino acids with role in inhibitor binding are highlighted: Inhibitor-2 binding amino acids (1.-7. blue squares); microcystin binding amino acids (1.-4. orange squares); okadaic acid and microcystin binding amino acids (1.-5. green squares); inhibitor-2, microcystin and okadaic acid binding amino acid (1.-2. blue boxes). A. thaliana type 1 protein phosphatase (TOPP1) was included in the alignment as marker for the conserved STP residues. The NCBI/GenBank identifiers of the sequences used to construct the alignment can be found in Table S1.
(B) High resolution 2D projection of SHLP1-GFP asexual blood stage and gametocyte co-stained with ER tracker. Bar = 10 μM.