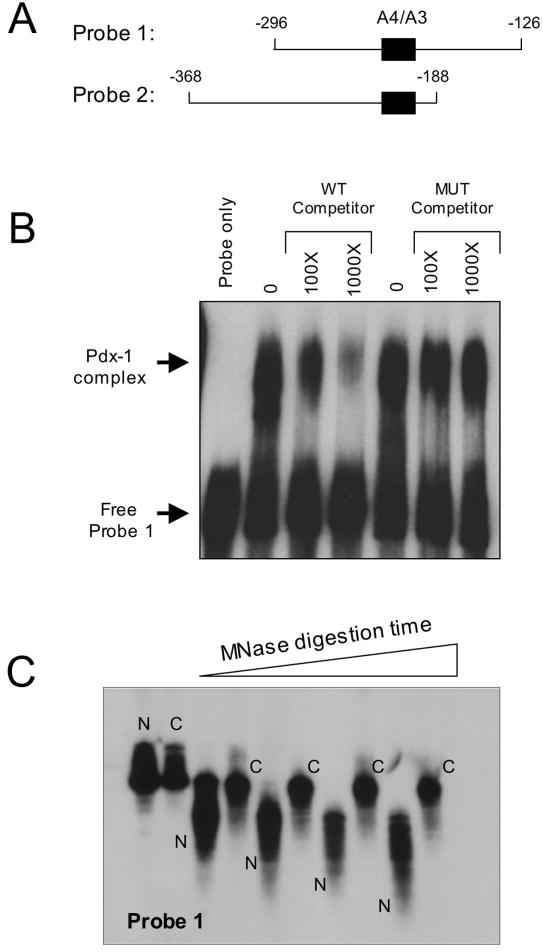
Figure 2. Binding of Pdx-1 to the insulin A4/A3 element and chromatin reconstitution of insulin gene fragments.
A, schematic representation of the two insulin probes used in chromatin reconstitution studies. The location of the A4/A3 element is indicated by the black box, and the numbers indicate positions of the sequences relative to the transcriptional start site of the insulin 1 gene. B, EMSA was performed using 20 ng bacterially-purified His6-Pdx-1 and approximately 32P-labeled Probe 1. Approximate molar excess of unlabeled competitor oligonucleotide is indicated. The mutant (MUT) competitor contains single bp mutations that disrupt the TAAT consensus sequences, as indicated in Materials and Methods. C, 32P-labeled Probe 1 was reconstituted as chromatin using a salt dilution technique as indicated in Materials and Methods. Naked (N) and chromatinized (C) probes were incubated with MNase for varying times (0-10 min), then treated with proteinase K to digest protein, then subjected to electrophoresis on a 5% polyacrylamide gel and to autoradiography.