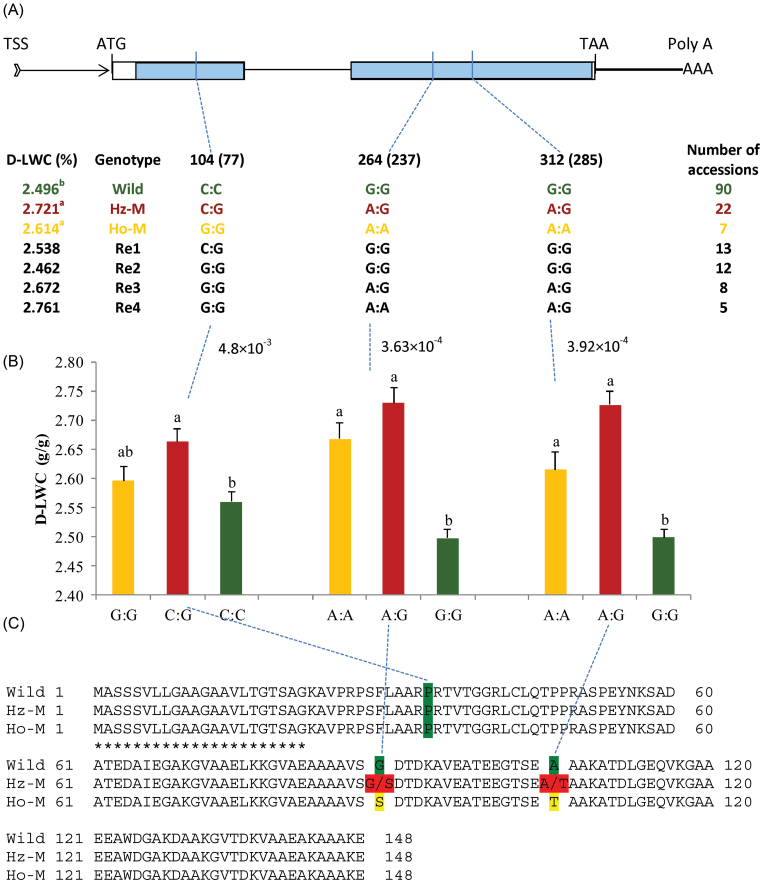
Fig. 4.
Variant alleles and association mapping at LpLEA3. (A) Gene structure and genotypes analysis of LpLEA3: wild type, heterozygous mutation (Hz-M), homozygous mutation (Ho-M), and recombinants one to four (R1–R4) genotypes were identified on the basis of LpLEA3 sequence from 158 perennial ryegrass accessions. The positions of these three significant SNPs were shown using the full-length LpLEA3 gene sequence as reference, with the start codon designated as position zero. The SNP positions at 104, 264, and 312 were located according to the reference mentioned above, whereas 77, 237, and 285 (in parentheses) were SNP positions relative to the SNP calling reference (accession 120 sequence). The proposed wild-type genotype was highlighted in green, Hz-M in red, Ho-M in yellow, and R1–R4 in black, respectively. Values of leaf water content under drought conditions (D-LWC) with the same letter were not significantly different at P < 0.05 for the wild type, Hz-M, and Ho-M. TSS, transcription start site; arrowed band, untranslated region; thick bar, downstream region after the stop codon; blank box, exon; thin line, intron; ATG, start codon; TAA, stop codon; blue shaded area, region sequenced in this study. (B) Leaf water content under drought stress (D-LWC) against its significant associated SNP markers. Values beside dashed blue lines indicate P-values of association mapping results. The wild-type genotype was highlighted in green, Hz-M in red, and Ho-M in yellow, respectively. Values of D-LWC with the same letter were not significantly different at P < 0.05. (C) Amino acid substitutions in LpLEA3 among wild type, Hz-M, and Ho-M. Underlining with 21 asterisks indicate its signal peptide.