Abstract
High levels of genetic diversity in Plasmodium falciparum populations are an obstacle to malaria control. Here, we investigate the relationship between local variation in malaria epidemiology and parasite genetic diversity in Papua New Guinea (PNG). Cross-sectional malaria surveys were performed in 14 villages spanning four distinct malaria-endemic areas on the north coast, including one area that was sampled during the dry season. High-resolution msp2 genotyping of 2,147 blood samples identified 761 P. falciparum infections containing a total of 1,392 clones whose genotypes were used to measure genetic diversity. Considerable variability in infection prevalence and mean multiplicity of infection was observed at all of the study sites, with the area sampled during the dry season showing particularly striking local variability. Genetic diversity was strongly associated with multiplicity of infection but not with infection prevalence. In highly endemic areas, differences in infection prevalence may not translate into a decrease in parasite population diversity.
Introduction
Plasmodium falciparum, the most virulent of the human malaria parasites, is the primary target of malaria control and elimination programs currently being rolled out around the world. There has been an increasing awareness that to sustain any reductions in the burden of malaria there is a need to understand the complex changes in malaria epidemiology that occur as a result of intensifying control efforts, particularly in regions where the disease is endemic.1 Molecular epidemiology can be used to monitor the impact of interventions by providing a sensitive and specific measure of not only the prevalence of infection, but also the multiplicity of infection (MOI). The MOI is measured by determining the number of genetically distinct parasite clones infecting individuals, and is associated with the level of transmission.2 Another less well-recognized but nevertheless important molecular parameter to consider is the genetic diversity of a parasite population because it is an indication of evolutionary fitness.3 On a global scale, the genetic diversity of P. falciparum is strongly associated with distance from Africa where P. falciparum malaria is thought to have originated.4 It has also been widely reported that diversity is associated with the overall level of transmission in the different continental regions.2,5,6 However, within these regions, fine-scale variations in transmission occur as a result of factors such as altitude, vector availability, urbanization, and malaria control, and these correspond with differences in genetic diversity.7–11 Diversity underlies resilience against interventions that directly target the parasite, such as malaria vaccines, which will need to cover all antigenically distinct strains circulating in an endemic area to be effective,12 and the emergence of resistance to antimalarial drugs.13 In the context of control programs, a diverse parasite population maintained in the face of falling prevalence would continue to be a significant public health challenge.
Papua New Guinea (PNG) has the highest burden of malaria in the Pacific region and outside Africa, with intense year-round transmission of all four of the major human malaria parasites (P. falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale s.l.) in the low-lying and coastal regions.14,15 The P. falciparum population is genetically complex with genotyping of antigens (msp1 and msp2) and putatively neutral microsatellite haplotypes revealing moderate to high levels of genetic diversity.2,16–18 We have previously investigated the genetic diversity of 318 monoclonal P. falciparum isolates from 14 villages spread over four distinct catchment areas of the Madang and East Sepik Provinces of PNG, by genotyping 10 microsatellite markers. These surveys revealed high levels of genetic diversity throughout the study area. Diversity was not associated with the P. falciparum infection prevalence, but lower levels of diversity were found in the more isolated inland populations.18
Since 2003, efforts to control malaria in PNG have been significantly intensified with support from the Global Fund to Combat AIDS, Tuberculosis, and Malaria. The strategies being used have included the extensive distribution of long-lasting insecticide-treated nets (LLINs) and more recently, policy change to ensure better diagnosis of infections and the treatment of clinical cases with artemisinin combination therapy.14 To further investigate the relationship between parasite genetic diversity and the endemicity of P. falciparum in PNG, we have analyzed the 14 P. falciparum populations by genotyping all P. falciparum infections (N = 761) at the highly polymorphic msp2 locus. Capillary electrophoresis allowed high-resolution detection of infection prevalence and MOI19,20 and the genetic diversity of each population. The results highlight the potential challenges of malaria control in highly endemic areas and the importance of molecular monitoring of malaria parasite populations during the implementation of control measures.
Materials and Methods
Study area.
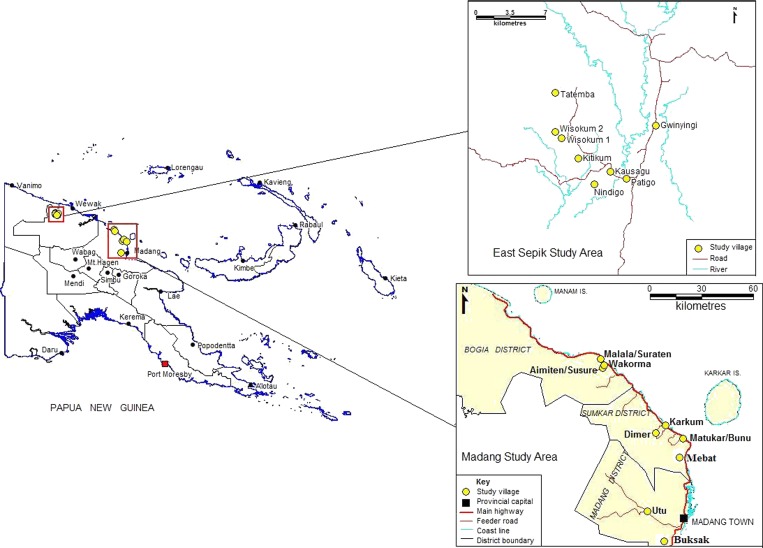
The study was conducted in the East Sepik and Madang Provinces on the north coast of PNG (Figure 1), where malaria transmission is intense all year round. In Madang Province, the entomological inoculation rate is 0.19–1.44 infective bites/person/night21 with a P. falciparum infection prevalence of 33% by light microscopy (LM)22 and multiple clones observed in 40–50% of infections.2,23 Although traditionally considered holoendemic,24,25 more recently malaria transmission in the Wosera region of the East Sepik Province has dropped, ranging between 0.15 and 0.7 infective bites/person/night26–28 with an infection prevalence of 19.5–23.9%29 and multiple infections are observed in 26% of infections,29 below that observed in many other parts of Madang and East Sepik Province.15
Figure 1.
Map of the study area.
The study sites in Madang included 12 villages distributed among three catchment areas—Mugil: comprising Matukar, Bunu, Karkum, and Dimer; Malala: comprising Wakorma, Malala, Suraten, Amiten, and Susure; and Utu: comprising three villages around the region around the village of Utu Sub-Health Center. Although Mugil and Malala are located primarily on the coast, Utu is found inland to the west of Madang in the Transgogol river region (Figure 1). There is a direct road between the catchments of Mugil and Malala, 50 km to the north of Madang town. Road access to Utu is to the west of Madang town along an unsealed road. Because of their close proximity three village pairs were compared as one village, namely Matukar-Bunu, Malala-Suraten, and Amiten-Susure. No specific location information was available for the Utu villages so Utu was analyzed as a catchment and village (Figure 1). Therefore, a total of seven parasite populations were analyzed for Madang Province.
The study sites in the East Sepik region included seven villages located within a 10 km radius in the Wosera-Gawi district, ∼75 km WSW of the provincial capital Wewak and 15–20 km S of the local administration center in Maprik (Figure 1). The villages included Gwinyingi in the north east, Patigo, Kausaugu, and Nindigo in the south, and Kitikum, Wisogum, and Tatemba in the north east (Figure 1).
Study population and samples.
Cross-sectional surveys were conducted in Madang Province in the rainy season period of March 2006 and in the East Sepik Province in the relatively dry months of August and September of 2005. These Madang surveys were done before a large-scale distribution of LLIN, whereas the Maprik villages had seen a LLIN mass distribution in the preceding 12 months.30 Although the incidence of P. falciparum malaria in PNG does show significant seasonal variation,25,31 the overall prevalence of P. falciparum is remarkably stable.29
Finger prick or venous blood samples were collected from asymptomatic individuals of all ages residing in the villages listed previously. The presence of parasites was diagnosed by LM and by polymerase chain reaction (PCR, see below). Genomic DNA was extracted from whole blood samples using the 96-well QiaQuick DNA extraction kit (Qiagen, Valencia, CA). Ethical approval to conduct the study was granted by the PNG Institute of Medical Research Institutional Review Board (0808), the Papua New Guinea Medical Research Advisory Committee (10.23), and the Alfred Hospital Research and Ethics Unit (30/06Q).
PCR and genotyping.
To identify and genotype P. falciparum positive samples, high-resolution msp2 genotyping was performed as previously described19; this method is specific to P. falciparum therefore only DNA samples positive for P. falciparum yield PCR products. Briefly, a nested multiplex PCR approach was used to amplify 3D7 and/or FC27 family alleles of the gene encoding the highly polymorphic antigen, msp2, using family-specific primers labeled with a fluorescent dye. The PCR products were analyzed by 1.5% agarose gel electrophoresis with PCR-positive samples being selected for further analysis. The allele family and fragment size were then determined by capillary electrophoresis using an ABI 3730×1 DNA Analyzer platform with the internal size standard GSLIZ500.
Analysis.
The number, family, and size of msp2 alleles were determined using Peak Scanner version 1.0 (Applied Biosystems, Carlsbad, CA). From these results, the PCR-based infection prevalence and MOI were defined. The P. falciparum parasite rate (PfPR) was measured as the proportion of samples that were infection positive by LM and by msp2 PCR; and the MOI was determined by counting the number of msp2 alleles detected within a sample. Both PCR and LM-based PfPR measures were adjusted for comparison across populations using the method of Smith and others32, which calculates values on the basis of PfPR in 2–10-year-old children (PfPR2–10). This is the same method as that used in the Malaria Atlas Project to map malaria endemicity worldwide, however only LM values have been used previously.33
Population genetic analyses were done using FSTAT software version 2.9.334 to define allele frequencies, the number of alleles (A), the allelic richness (Rs), which is a normalized measure of the number of alleles35 and the expected heterozygosity, calculated as
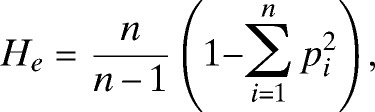 |
where p is the frequency of the ith allele and n is the number of alleles in the sample. Statistical analysis was done using nonparametric methods including Mann Whitney U tests to detect a difference in values and Spearman's Rho test to measure associations, unless otherwise stated. Analysis was done using the software package, SPSS v17 (SPSS, Inc., Chicago, IL).
Results
A total of 2,308 volunteers from 14 villages or village pairs and four distinct catchment areas (Mugil, Malala, Utu, and Wosera) were included in the study (Figure 1). Volunteers included 770 (33.4%) children < 10 years of age and 889 (38.5%) adults > 20 years of age. Key demographic data used in the study (age or village) were missing for 85 samples. Of 2,205 samples (95.5%) screened by msp2 PCR, a total of 578 (26.2%) samples contained P. falciparum parasites by LM and 766 (34.8%) by PCR. The msp2 allele sizes were unable to be determined for five samples, thus msp2 alleles were defined for 761 (34.5%) P. falciparum isolates.
Spatially variable prevalence and multiplicity of P. falciparum infections in Papua New Guinea.
PCR-based species typing methods have recently shown higher infection prevalence and a later age-peak than that estimated by LM.15 The msp2 PCR in this new sample set also confirmed a significantly higher prevalence of P. falciparum infection by PCR compared with LM, with a 1.1- to 2.3-fold higher infection prevalence (paired t test: P = 0.0004, Table 1) however the results for the two methods were strongly correlated (Spearman's Rho test: ρ = 0.81, P = 0.0004). The East Sepik village of Kausagu was an exception however, with lower infection prevalence by PCR than by LM. This may be explained by the fact that a larger number of samples were screened by LM (N = 75) than by PCR (N = 58), or it may be a result of false positive diagnosis by LM. The catchment of Mugil had the highest infection prevalence and least variability among villages (by both LM and PCR) followed by Utu and Malala (Table 1). However, Wosera, where more villages were surveyed, had substantial variability in the infection prevalence. The highest (Nindigo = 43.5% by PCR) and lowest (Kausagu = 1.7% by PCR) infection prevalence rates in the entire study area were found in this area, which is surprising because the distance between them is only ∼2 km (Table 1, Figure 1). Overall, the villages in Madang had a higher prevalence of infection than those in the East Sepik as measured by LM (Mann Whitney U test: P = 0.015) but not by PCR (P = 0.14).
Table 1.
Intensity of Plasmodium falciparum transmission in 14 villages of Papua New Guinea*
| Province | Catchment | n2–10 | PfPR2–10 | Mean multiplicity of infection (range) | ||
|---|---|---|---|---|---|---|
| Village | ntotal | LM | PCR | |||
| MADANG | MUGIL | 493 | 170 | 38.1 | 55.7 | 2.01 (1–11) |
| Matukar/Bunu | 210 | 77 | 41.8 | 56.2 | 2.12 (1–11) | |
| Karkum | 209 | 59 | 39.4 | 55.2 | 2.07 (1–10) | |
| Dimer | 74 | 34 | 45.9 | 55 | 1.53 (1–4) | |
| MALALA | 387 | 131 | 35.5 | 44.6 | 1.59 (1–8) | |
| Wakorma | 150 | 48 | 38.1 | 45.9 | 1.44 (1–4) | |
| Malala/Suraten | 65 | 22 | 32.8 | 44.6 | 1.50 (1–8) | |
| Amiten/Susure | 172 | 61 | 33.5 | 42.1 | 1.77 (1–5) | |
| UTU | 395 | 127 | 45.2 | 49.7 | 1.97 (1–13) | |
| Utu | 395 | 127 | 45.2 | 49.7 | 1.97 (1–13) | |
| EAST SEPIK | WOSERA | 930 | 304 | 24.9 | 36.1 | 1.72 (−6) |
| Gwinyingi | 119 | 39 | 22.9 | 48.7 | 1.69 (1–6) | |
| Patigo | 113 | 50 | 6.9 | 16.2 | 1.00 (1–1) | |
| Kausagu | 58† | 20 | 9.1 | < 1 | 2.00 (2–2) | |
| Nindigo | 216 | 55 | 41.9 | 56.2 | 1.66 (1–5) | |
| Kitikum | 124‡ | 55 | 28.4 | 47.8 | 2.08 (1–5) | |
| Wisokum | 200 | 55 | 16.3 | 22.2 | 1.62 (1–5) | |
| Tatemba | 100 | 30 | 29.6 | 32.2 | 1.84 (1–5) | |
ntotal = total number of samples screened; n2–10 = number of 2–10-year-old children screened; PfPR2–10 = infection prevalence (%) among 2–10-year-old children; LM = light microscopy; PCR = polymerase chain reaction msp2 genotyping.
of 75 samples,
of 200 samples.
The MOI was also variable throughout the study area, with mean MOI ranging from solely single infections in Patigo (Wosera) to a mean of 2.12 P. falciparum clones per sample in Matukar/Bunu (Mugil). The LM and PCR prevalence were not correlated with MOI (ρ = 0.143 and 0.323, respectively, P > 0.26) even when the outlier, Kausagu, was removed (ρ = 0.247, 0.487, respectively, P > 0.091) (Table 1). Overall, there was no significant difference in the multiplicity of infection in the East Sepik and Madang Provinces (Mann Whitney U test: P = 1.0).
These data show that the overall P. falciparum endemicity based on PCR-based measures of infection prevalence, and the MOI, are similar between the provinces of Madang and East Sepik, despite the latter province being sampled in the dry season and following a LLIN distribution. When individual catchments and villages were compared, variability was observed both within Madang, with a higher prevalence and MOI observed in the Mugil villages followed by Utu and Malala, and within the Wosera, which had the lowest prevalence overall and considerable variability among the villages. Therefore, prevalence among villages in each of the Madang catchments surveyed during the rainy months and pre-LLIN was relatively uniform; however, in the Wosera catchment surveyed during the drier months and post-LLIN distribution, there were a number of “infection hotspots” interspersed with relatively low transmission villages.
Genetic diversity of P. falciparum in Papua New Guinea.
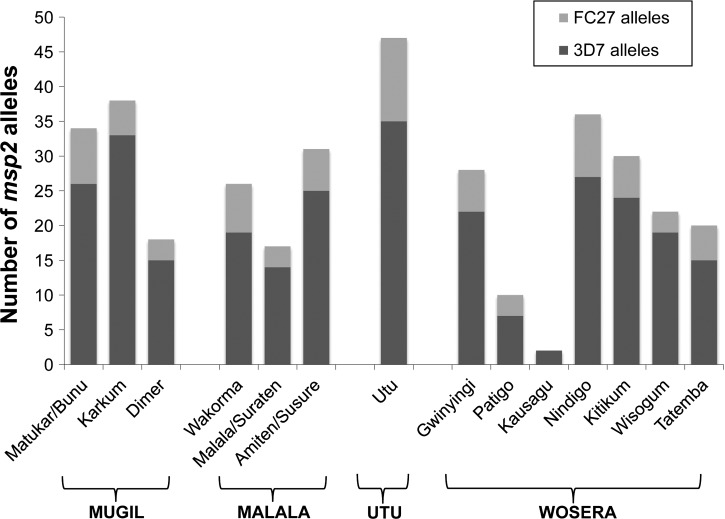
Among the 761 P. falciparum PCR-positive samples that were successfully genotyped, 1,392 distinct parasite clones were identified. Of these, 68% of clones harbored 3D7-type msp2 alleles and 32% had FC27-type msp2 alleles. Among these clones, there were 83 distinct msp2 alleles including 63 3D7-type alleles ranging in size 171–485 bp and 20 FC27-type alleles ranging in size 184–512 bp (Supplemental Table S1). This confirms a high degree of sensitivity of the typing method to detect multiple infections in PNG.36 Similar proportions of 3D7 and FC27-type alleles were present in each population (Supplemental Table S1, Figure 2).
Figure 2.
Pfmsp2 allele-family distribution in 14 Plasmodium falciparum populations of Papua New Guinea. The number of Pfmsp2 alleles and proportion of the two allele-families (3D7 and FC27) are shown for each village.
In population genetics, large sample sizes are desirable to ensure that the majority of the diversity has been defined. To determine the optimal sample size for future surveys, we examined the number of alleles as a function of the number of clones sampled. On the basis of this information, we predict that 150 clones (∼100 P. falciparum isolates) are required to sample the majority of the msp2 alleles in PNG populations (Supplemental Figure S1). The lower prevalence of infection in some Wosera villages (e.g., Patigo, Kausagu) resulted in a smaller sample size than this and therefore the full extent of the diversity in these villages has not been defined. As a result, the number of alleles (A) detected was strongly associated with the number of P. falciparum isolates genotyped (Table 2; ρ = 0.974, P < 0.0001). However, the allelic richness (Rs), which is normalized on the basis of the smallest sample size, was also associated with the sample size (Table 2; ρ = 0.720, P = 0.006) suggesting that parasite diversity is associated with the number of infections in a given area. The expected Heterozygosity (He), a combined measure of the number of different alleles and their frequencies, was not associated with sample size (Table 2; ρ = −0.138, P = 0.654), though there was very little variability among the He values with all villages showing extremely high values levels of diversity based on this statistic (see below).
Table 2.
Genetic diversity of Plasmodium falciparum in 14 villages of Papua New Guinea*
| Catchment | c | A | Rs | He | |
|---|---|---|---|---|---|
| Village | n | ||||
| MUGIL | 211 | 424 | 47 | 43.4 | 0.93 |
| Matukar/Bunu | 91 | 193 | 34 | 14.21 | 0.92 |
| Karkum | 88 | 182 | 38 | 15.44 | 0.94 |
| Dimer | 32 | 49 | 18 | 12.91 | 0.93 |
| MALALA | 133 | 211 | 44 | 44.0 | 0.92 |
| Wakorma | 55 | 79 | 26 | 13.59 | 0.90 |
| Malala/Suraten | 22 | 33 | 17 | 13.25 | 0.92 |
| Amiten/Susure | 56 | 99 | 31 | 14.97 | 0.93 |
| UTU | 156 | 308 | 47 | 45.4 | 0.95 |
| Utu | 156 | 308 | 47 | 16.54 | 0.95 |
| WOSERA | 261 | 449 | 58 | 51.3 | 0.94 |
| Gwinigi | 45 | 76 | 28 | 15.37 | 0.95 |
| Patigo | 14 | 14 | 10 | 10.00 | 0.96 |
| Kausagu | 1 | 2 | 2 | n.d. | n.d |
| Nindigo | 94 | 156 | 36 | 14.11 | 0.92 |
| Kitikum | 48 | 100 | 30 | 15.03 | 0.94 |
| Wisogum | 34 | 55 | 22 | 13.80 | 0.93 |
| Tatemba | 25 | 46 | 20 | 13.81 | 0.93 |
n = number of P. falciparum isolates; c = number of P. falciparum clones; A = number of alleles; Rs = allelic richness; He = expected heterozygosity.
The genetic diversity of villages throughout the study area, based on Rs and He, was high and variable as previously shown using microsatellite haplotypes.18 Utu, the Mugil village of Karkum, and Wosera villages of Gwinyingi and Kitikum had the highest values of Rs, and the Wosera village of Patigo the lowest. There was a limited variability of He among the villages, with all showing very high levels of diversity (He = 0.90–0.96). Patigo had the highest He, probably as a result of the smaller sample size resulting in an overestimate of diversity (Table 2).
The msp2 diversity data were then compared with that previously published for microsatellite haplotypes.18 Microsatellite haplotype diversity (Rs) was much lower than that for msp2 (Supplemental Figure S2). In this study a higher resolution marker, such as msp2 was necessary to allow the distinction between clones within samples.
Association between endemicity and genetic diversity.
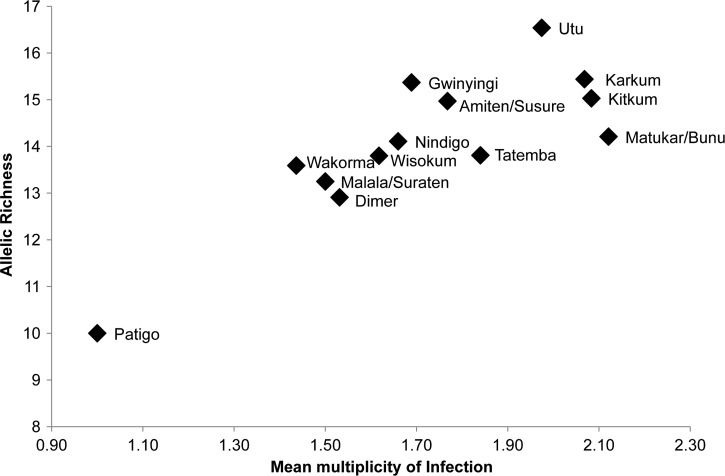
The variability in prevalence and MOI among the villages, especially within the Wosera area provided the opportunity to investigate associations with msp2 diversity, as defined by the unbiased estimates of Rs and He. Neither LM nor PCR-based prevalence measures were associated with Rs (ρ = 0.203, 0.448, respectively, P > 0.125) or He (ρ = −0.356, −0.213, P > 0.230). As expected, the mean MOI was significantly associated with Rs (ρ = 0.797, P = 0.001; Figure 3) and remained significant when the outlier Patigo was removed (ρ = 0.741, P = 0.005). However, there was no significant association of MOI with He values (ρ = 0.152, P = 0.621) most likely because all He values were so close to the maximum value of 1. A strong association was observed between the allelic richness (Rs) and the number of P. falciparum clones observed in each population (ρ = 0.791, P = 0.001) confirming that populations with the largest numbers of clones are also the most diverse.
Figure 3.
Relationship between multiplicity of infection and Plasmodium falciparum genetic diversity in Papua New Guinea. Genetic diversity was measured by determining the Pfmsp2 allelic richness (Rs) for each village and plotted against the mean multiplicity of infection (MOI), which is based on the number of clones per parasite isolate.
Discussion
For the first time in decades real progress is being made in the global control of malaria. Gains are being made in the heartland of high-intensity transmission and a strong malaria elimination agenda is emerging in those areas that have already reduced transmission to low levels.1 This increasing success has highlighted a number of gaps in our knowledge of the complex and dynamic interplay of endemicity, immunity, and parasite diversity. Fundamental questions about the effect of decreased malaria transmission on the parasite and the host need answering, to use the most effective interventions and to be able to predict the consequences and protect the human population in the face of falling immunity. A very basic question is the impact of endemicity on parasite diversity on a local scale. This study has confirmed and extended previous reports based on LM-based detection of parasites that the endemicity of P. falciparum malaria in PNG is heterogeneous on both a national and local scale.24,37 High-resolution genotyping of the highly polymorphic marker, msp2 allowed the measurement of the infection prevalence (PfPR) and the MOI, which were defined for 14 villages spread over four catchments in the provinces of Madang (N = 3 catchments; 7 villages) and East Sepik (N = 1 catchment, 7 villages). As previously shown in PNG,15,29 the nested PCR approach was effective at identifying sub-microscopic infections and clones, and provided up to twofold higher sensitivity than LM in diagnosing infection with P. falciparum, though the relative levels of infection prevalence among populations were concordant with those determined by LM. The detection of 83 distinct msp2 alleles throughout the study area confirms high levels of sensitivity of the methodology to detect multiple clones.36 The molecular data also provided the opportunity to measure the genetic diversity in villages with a wide range of infection prevalences, particularly those within the East Sepik region during the dry season and post-LLIN distribution.
The infection prevalence based on molecular approaches to define PfPR2–10, and the mean MOI was highest in the catchment of Mugil followed by Utu, and lowest in Wosera and Malala, respectively. In Madang Province, individual villages of Mugil also showed the highest values for these parameters and Malala the lowest. However in the East Sepik, villages within the single catchment of the Wosera were highly variable showing the widest range of PfPR2–10 and MOI values for the entire study area. The East Sepik Province has experienced a significant reduction in P. falciparum prevalence of 38% in the early 1990s down to 22.3% in 2002.24,29 Our data show that P. falciparum prevalence in this region remained relatively stable between August and November of 2002 (19.5%)29 and the same time period in 2005 (20.1%) when the samples used in the current study were collected. A further significant decline in infection prevalence in the Wosera has been noted in recent years and is thought to be the result of ongoing malaria studies in the region for more than 20 years leading to greater treatment and awareness, and has resulted in almost universal bednet use. Data collected during our surveys shows that bednets were being used by > 97.1% of study volunteers in the Wosera and Malala villages, whereas 95% reported bednet use for Utu and 25.7%, 60.1%, and 96.6% for the three Mugil villages (Mueller and others, unpublished data). However, although a distribution of LLINs took place in the Wosera in 2005, most bednets used in the Madang areas were neither impregnated nor long lasting.
The dominance and higher diversity of the msp2 3D7 allele family throughout PNG has important implications for vaccine trials in the region. A subunit vaccine formulated with this msp2 allele that produced cross-reactive responses against other 3D7 alleles would have a greater chance of success than would that containing a FC27 allele. Indeed, one of the most successful vaccine trials to date included a 3D7 allele of msp2 and showed a selective effect against infection with parasites harboring this genotype.38 Interestingly, previous surveys of msp2 have shown that in 1992 the FC27 allele was at almost equal frequencies with 3D7,39 whereas more recent surveys in the Wosera region have confirmed the contemporary dominance of 3D7 in PNG.20 Nevertheless, a successful msp2-based malaria vaccine would likely need to contain both 3D7 and FC27 alleles.
In the PNG populations surveyed, the genetic diversity was less variable than the prevalence and the MOI. A narrow range for both diversity statistics (He and Rs) suggests that diversity is maintained at high levels despite the different levels of malaria endemicity in the catchments. High diversity is also found during the drier months in the Wosera. This is consistent with the lack of seasonality in the MOI of P. falciparum infections in a cohort of young children in a neighboring population (Mueller and others, unpublished data). The genetic diversity of a parasite population is shaped by various influences, including genetic drift, mutation, natural selection, and gene flow.40 MSP2 is the second most abundant protein on the surface of the merozoite41 and a target of naturally acquired antibodies,42,43 and therefore diversity is influenced by immune (balancing) selection. A comparison of msp2 diversity between Tanzania and PNG showed many more alleles (A = 76 and 35, respectively), however only slightly higher expected heterozygosity (He = 0.933 and 0.965, respectively) in the African population where transmission is approximately ten times that of PNG.20 Therefore, it is important to consider that the diversity seen in msp2 reflects both immune selection and the intensity of transmission. The lack of correlation between diversity and infection prevalence suggests that in the PNG populations, diversity may have reached a maximal level and it will only be through substantial and sustained reductions in transmission that a decrease in diversity will be possible.
One explanation for the high levels of diversity in all populations of the Wosera is that parasite populations in the region are panmictic. However, surveys of the same populations using a panel of 10 microsatellite markers have demonstrated that the catchments and even some villages within the same catchment are genetically isolated from each other, suggesting that there is a limit to the influx of alleles from distant and even nearby populations.18 Diversity as measured by these putatively neutral markers was also high and there was no correlation with the infection prevalence.18 It may be that the reduction of transmission in the dry season and by the recent distribution of LLINs in the Wosera has created a situation in which diverse populations have become isolated. To confirm this, it will be important to later compare the data to that for the rainy season and pre-LLIN distributions.
Though the diversity of msp2 was not influenced by the lower prevalence, the allelic richness was strongly correlated with the MOI, consistent with the requirement for transmission of multiple clones to create new genotypes within the mosquito midgut.23,44 Studies in western Kenya, before and 5 years after insecticide-treated nets were introduced have shown that despite drastically decreased prevalence, the proportion of multiple infections did not change.45 The Kenyan study also demonstrated using eight microsatellite markers, that diversity (as measured by the expected heterozygosity) remained high overall and for the majority of markers.45 Studies in the Peruvian Amazon have shown that the MOI actually increases with decreasing prevalence, which will ultimately maintain population complexity.46 Together with the findings reported here, this suggests that the diversity of parasite populations and therefore the resilience to antimalarial interventions will remain high unless the MOI can be significantly reduced. A better understanding of the relationship between parasite prevalence, multiplicity of infection, and genetic diversity is needed in the context of intensifying malaria control.
This study presents the molecular epidemiology of P. falciparum in the early stages of the intensification of malaria control in PNG. We have demonstrated by high-resolution genotyping of P. falciparum isolates from 14 villages spread across four distinct catchment areas of PNG, that the prevalence of P. falciparum malaria is highly variable among villages, and most notably so in the Wosera region of the East Sepik Province after a recent LLIN distribution. Despite this variability, the genetic diversity in all populations was at similarly high levels throughout the study area. Furthermore, although the variable prevalence in the East Sepik Province (Wosera catchment) indicates a recent decline in transmission intensity in some villages (caused by seasonal fluctuations, better access to efficient antimalarial treatment, and recent near-universal bednet use), parasite genetic diversity was similar to that in three catchments of Madang Province. High levels of diversity indicate a malaria population with a greater capacity to evade interventions and thus more difficult to control. Molecular typing may therefore provide deeper insight into the longer-term impact of interventions, in addition to the infection prevalence, which has traditionally been monitored by LM. This study highlights the use of molecular epidemiology to monitor changes in the structure of parasite populations and warrants continued investigations in the context of intensifying malaria control efforts in PNG.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the ongoing and generous support of the Papua New Guinean communities, in particular the volunteers and their families and the staff of the Papua New Guinea Institute of Medical Research, the study would not have been possible without this support. We also extend our thanks to O. Toporua (PNGIMR) and J. Wapling (Burnet Institute) for technical support; E. Namosha for providing maps of the study area (PNGIMR); T. Adiguma (PNGIMR) for furnishing appropriate sample information; and P. Gething and S. Hay for calculating PfPR2–10 from the data sets.
Footnotes
Financial support: The samples used in the study were collected during a study of Intermittent Preventive Treatment in infants (IPTi) for malaria in Papua New Guinea supported by the Bill & Melinda Gates Foundation. This work was made possible with the support of the National Health and Medical Research Council (NHMRC) of Australia through project grant 488221 and a Research Fellowship awarded to JCR, the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
Authors' addresses: Alyssa E. Barry, Division of Infection and Immunity, Walter and Eliza Hall Institute for Medical Research, Victoria, Australia, E-mail: barry@wehi.edu.au. Lee Schultz, BioSciences Research Division, Department of Primary Industries, Victoria, Australia, E-mail: Lee.Schultz@dpi.vic.gov.au. Nicolas Senn, Swiss Tropical and Public Health Institute, Basel, Switzerland, E-mail: Nicolas.senn@gmail.com. Joe Nale, Benson Kiniboro, and Peter M. Siba, Papua New Guinea Institute of Medical Research, Papua New Guinea, E-mails: jnalejr@gmail.com, benson.kiniboro@pngimr.org.pg, and Peter.Siba@pngimr.org.pg. Ivo Mueller, Division of Infection and Immunity, Walter and Eliza Hall Institute for Medical Research, Victoria, Australia, E-mail: mueller@wehi.edu.au. John C. Reeder, Centre for Population Health, Burnet Institute, Victoria, Australia, E-mail: jreeder@burnet.edu.au.
References
- 1.Feachem RG, Phillips AA, Targett GA, Snow RW. Call to action: priorities for malaria elimination. Lancet. 2010;376:1517–1521. doi: 10.1016/S0140-6736(10)61500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 3.DHaFer R. Correlation between fitness and genetic diversity. Conservation Ecology. 2003;17:231–237. [Google Scholar]
- 4.Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, Ranford-Cartwright L, Sawai H, Sakihama N, Ohmae H, Nakamura M, Ferreira MU, Escalante AA, Prugnolle F, Bjorkman A, Farnert A, Kaneko A, Horii T, Manica A, Kishino H, Balloux F. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 5.Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 7.Anthony TG, Conway DJ, Cox-Singh J, Matusop A, Ratnam S, Shamsul S, Singh B. Fragmented population structure of Plasmodium falciparum in a region of declining endemicity. J Infect Dis. 2005;191:1558–1564. doi: 10.1086/429338. [DOI] [PubMed] [Google Scholar]
- 8.Bogreau H, Renaud F, Bouchiba H, Durand P, Assi SB, Henry MC, Garnotel E, Pradines B, Fusai T, Wade B, Adehossi E, Parola P, Kamil MA, Puijalon O, Rogier C. Genetic diversity and structure of African Plasmodium falciparum populations in urban and rural areas. Am J Trop Med Hyg. 2006;74:953–959. [PubMed] [Google Scholar]
- 9.Durand P, Michalakis Y, Cestier S, Oury B, Leclerc MC, Tibayrenc M, Renaud F. Significant linkage disequilibrium and high genetic diversity in a population of Plasmodium falciparum from an area (Republic of the Congo) highly endemic for malaria. Am J Trop Med Hyg. 2003;68:345–349. [PubMed] [Google Scholar]
- 10.Leclerc MC, Durand P, de Meeus T, Robert V, Renaud F. Genetic diversity and population structure of Plasmodium falciparum isolates from Dakar, Senegal, investigated from microsatellite and antigen determinant loci. Microbes Infect. 2002;4:685–692. doi: 10.1016/s1286-4579(02)01587-3. [DOI] [PubMed] [Google Scholar]
- 11.Tsumori Y, Ndounga M, Sunahara T, Hayashida N, Inoue M, Nakazawa S, Casimiro P, Isozumi R, Uemura H, Tanabe K, Kaneko O, Culleton R. Plasmodium falciparum: differential selection of drug resistance alleles in contiguous urban and peri-urban areas of Brazzaville, Republic of Congo. PLoS ONE. 2011;6:e23430. [Google Scholar]
- 12.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, McNamara DT, Bockarie MJ, Kazura JW, Kyle DE, Fidock DA, Zimmerman PA. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hetzel MW. An integrated approach to malaria control in Papua New Guinea. P N G Med J. 2009;52:1–7. [PubMed] [Google Scholar]
- 15.Mueller I, Widmer S, Daniela M, Maraga S, McNamara DT, Kiniboro B, Sie A, Smith TA, Zimmerman PA. High sensitivity detection of Plasmodium species reveals positive correlations between infections of different species, shifts in age distribution and reduced local variation in Papua New Guinea. Malar J. 2009;8:41–55. doi: 10.1186/1475-2875-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, Walliker D, Day KP. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121:257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 17.Cortes A, Felger I, Beck HP. Molecular parasitology of malaria in Papua New Guinea. Trends Parasitol. 2003;19:246–249. doi: 10.1016/s1471-4922(03)00090-4. [DOI] [PubMed] [Google Scholar]
- 18.Schultz L, Wapling J, Mueller I, Ntsuke PO, Senn N, Nale J, Kiniboro B, Buckee CO, Tavul L, Siba PM, Reeder JC, Barry AE. Multilocus haplotypes reveal variable levels of diversity and population structure of Plasmodium falciparum in Papua New Guinea, a region of intense perennial transmission. Malar J. 2010;9:336. doi: 10.1186/1475-2875-9-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk N, Maire N, Sama W, Owusu-Agyei S, Smith T, Beck HP, Felger I. Comparison of PCR-RFLP and Genescan-based genotyping for analyzing infection dynamics of Plasmodium falciparum. Am J Trop Med Hyg. 2006;74:944–950. [PubMed] [Google Scholar]
- 20.Schoepflin S, Valsangiacomo F, Lin E, Kiniboro B, Mueller I, Felger I. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar J. 2009;8:250. doi: 10.1186/1475-2875-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkot TR, Graves PM, Paru R, Wirtz RA, Heywood PF. Human malaria transmission studies in the Anopheles punctulatus complex in Papua New Guinea: sporozoite rates, inoculation rates, and sporozoite densities. Am J Trop Med Hyg. 1988;39:135–144. doi: 10.4269/ajtmh.1988.39.135. [DOI] [PubMed] [Google Scholar]
- 22.Michon P, Cole-Tobian JL, Dabod E, Schoepflin S, Igu J, Susapu M, Tarongka N, Zimmerman PA, Reeder JC, Beeson JG, Schofield L, King CL, Mueller I. The risk of malarial infections and disease in Papua New Guinean children. Am J Trop Med Hyg. 2007;76:997–1008. [PMC free article] [PubMed] [Google Scholar]
- 23.Paul RE, Packer MJ, Walmsley M, Lagog M, Ranford-Cartwright LC, Paru R, Day KP. Mating patterns in malaria parasite populations of Papua New Guinea. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 24.Genton B, al-Yaman F, Beck HP, Hii J, Mellor S, Narara A, Gibson N, Smith T, Alpers MP. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I. Malariometric indices and immunity. Ann Trop Med Parasitol. 1995;89:359–376. doi: 10.1080/00034983.1995.11812965. [DOI] [PubMed] [Google Scholar]
- 25.Genton B, al-Yaman F, Beck HP, Hii J, Mellor S, Rare L, Ginny M, Smith T, Alpers MP. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. II. Mortality and morbidity. Ann Trop Med Parasitol. 1995;89:377–390. doi: 10.1080/00034983.1995.11812966. [DOI] [PubMed] [Google Scholar]
- 26.Hii JL, Smith T, Mai A, Ibam E, Alpers MP. Comparison between anopheline mosquitoes (Diptera: Culicidae) caught using different methods in a malaria endemic area of Papua New Guinea. Bull Entomol Res. 2000;90:211–219. doi: 10.1017/s000748530000033x. [DOI] [PubMed] [Google Scholar]
- 27.Bockarie MJ, Alexander N, Bockarie F, Ibam E, Barnish G, Alpers M. The late biting habit of parous Anopheles mosquitoes and pre-bedtime exposure of humans to infective female mosquitoes. Trans R Soc Trop Med Hyg. 1996;90:23–25. doi: 10.1016/s0035-9203(96)90465-4. [DOI] [PubMed] [Google Scholar]
- 28.Hii JL, Smith T, Vounatsou P, Alexander N, Mai A, Ibam E, Alpers MP. Area effects of bednet use in a malaria-endemic area in Papua New Guinea. Trans R Soc Trop Med Hyg. 2001;95:7–13. doi: 10.1016/s0035-9203(01)90315-3. [DOI] [PubMed] [Google Scholar]
- 29.Kasehagen LJ, Mueller I, McNamara DT, Bockarie MJ, Kiniboro B, Rare L, Lorry K, Kastens W, Reeder JC, Kazura JW, Zimmerman PA. Changing patterns of Plasmodium blood-stage infections in the Wosera region of Papua New Guinea monitored by light microscopy and high throughput PCR diagnosis. Am J Trop Med Hyg. 2006;75:588–596. [PMC free article] [PubMed] [Google Scholar]
- 30.Senn N, Rarau P, Stanisic DI, Robinson L, Barnadas C, Manong D, Salib M, Iga J, Tarongka N, Ley S, Rosanas-Urgell A, Aponte JJ, Zimmerman PA, Beeson JG, Schofield L, Siba P, Rogerson SJ, Reeder JC, Mueller I. Intermittent preventive treatment for malaria in Papua New Guinean infants exposed to Plasmodium falciparum and P. vivax: a randomized controlled trial. PLoS Med. 2012;9:e1001195. doi: 10.1371/journal.pmed.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin E, Kiniboro B, Gray L, Dobbie S, Robinson L, Laumaea A, Schopflin S, Stanisic D, Betuela I, Blood-Zikursh M, Siba P, Felger I, Schofield L, Zimmerman P, Mueller I. Differential patterns of infection and disease with P. falciparum and P. vivax in young Papua New Guinean children. PLoS ONE. 2010;5:e9047. doi: 10.1371/journal.pone.0009047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DL, Guerra CA, Snow RW, Hay SI. Standardizing estimates of the Plasmodium falciparum parasite rate. Malar J. 2007;6:131. doi: 10.1186/1475-2875-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malaria Atlas Project 2011. http://www.map.ox.ac.uk Available at.
- 34.Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 35.Hurlbert SH. The non-concept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 36.Liljander A, Wiklund L, Falk N, Kweku M, Martensson A, Felger I, Farnert A. Optimization and validation of multi-colored capillary electrophoresis for genotyping of Plasmodium falciparum merozoite surface proteins (msp1 and 2) Malar J. 2009;8:78. doi: 10.1186/1475-2875-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cattani JA, Tulloch JL, Vrbova H, Jolley D, Gibson FD, Moir JS, Heywood PF, Alpers MP, Stevenson A, Clancy R. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am J Trop Med Hyg. 1986;35:3–15. doi: 10.4269/ajtmh.1986.35.3. [DOI] [PubMed] [Google Scholar]
- 38.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, Rare L, Baisor M, Lorry K, Brown GV, Pye D, Irving DO, Smith TA, Beck HP, Alpers MP. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1-2b trial in Papua New Guinea. J Infect Dis. 2002;185:820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 39.Felger I, Tavul L, Kabintik S, Marshall V, Genton B, Alpers M, Beck HP. Plasmodium falciparum: extensive polymorphism in merozoite surface antigen 2 alleles in an area with endemic malaria in Papua New Guinea. Exp Parasitol. 1994;79:106–116. doi: 10.1006/expr.1994.1070. [DOI] [PubMed] [Google Scholar]
- 40.Hartl DL, Clark AG. Principles of Population Genetics. Sunderland, MD: Sinauer Associates; 1997. [Google Scholar]
- 41.Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, 3rd, Crabb BS. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 42.al-Yaman F, Genton B, Anders R, Taraika J, Ginny M, Mellor S, Alpers MP. Assessment of the role of the humoral response to Plasmodium falciparum MSP2 compared to RESA and SPf66 in protecting Papua New Guinean children from clinical malaria. Parasite Immunol. 1995;17:493–501. doi: 10.1111/j.1365-3024.1995.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 43.Stanisic DI, Richards JS, McCallum FJ, Michon P, King CL, Schoepflin S, Gilson PR, Murphy VJ, Anders RF, Mueller I, Beeson JG. Immunoglobulin G subclass-specific responses against Plasmodium falciparum merozoite antigens are associated with control of parasitemia and protection from symptomatic illness. Infect Immun. 2009;77:1165–1174. doi: 10.1128/IAI.01129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T, Walliker D. Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology. 1994;109:413–421. doi: 10.1017/s0031182000080665. [DOI] [PubMed] [Google Scholar]
- 45.Gatei W, Kariuki S, Hawley W, ter Kuile F, Terlouw D, Phillips-Howard P, Nahlen B, Gimnig J, Lindblade K, Walker E, Hamel M, Crawford S, Williamson J, Slutsker L, Shi YP. Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010;9:353. doi: 10.1186/1475-2875-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutton PL, Torres LP, Branch OH. Sexual recombination is a signature of a persisting malaria epidemic in Peru. Malar J. 2011;10:329. doi: 10.1186/1475-2875-10-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.