Figure 6. Structural and functional analyses of the Fbxl3-mCRY2 interface.
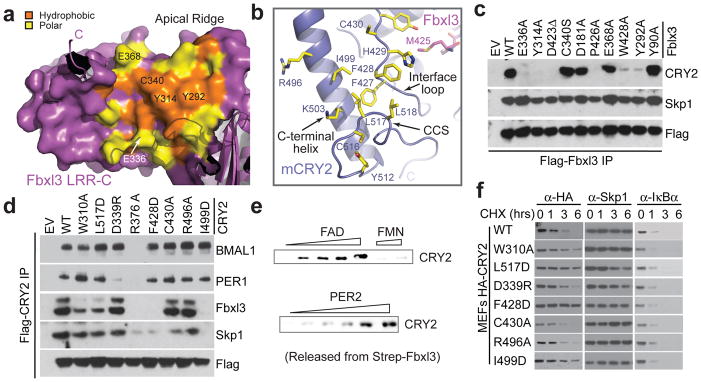
a, Surface and ribbon diagrams of the Fbxl3 LRR-C subdomain showing hydrophobic (orange) and polar (yellow) residues that are involved in mCRY2 binding. Amino acids selected for mutational analyses are labeled. The C-terminal tail and some N-terminal repeats are shown in ribbon diagrams. b, Ribbon diagrams of the mCRY2 structural elements involved in Fbxl3-LRR-C interaction. Select interface residues are shown in sticks. A part of the Fbxl3 tail (magenta) is shown for reference. c, Interactions of retrovirus-expressed Fbxl3 mutants with endogenous mCRY2 in mouse embryonic fibroblasts (MEF) assessed by co-immunoprecipitation and western blot analysis. d, Flag-mCRY2 complexes were immunoprecipitated with an anti-FLAG resin from transfected HEK293T cells and assessed for binding to Fbxl3, PER1, BMAL1 and Skp1. R376A served as a negative control as it is likely detrimental to mCRY2 folding. See Supplementary Discussion for C430A and Fbxl3-C340S mutants. e, Two in vitro competition assays demonstrating the ability of FAD and PER2, but not FMN, to disrupt a preformed Fbxl3-mCRY2 complex. f, MEFs were infected with retroviruses expressing either mCRY2 or mCRY2 mutants and treated with cycloheximide (CHX) for the indicated times. Total extracts were analyzed by immunoblotting.
