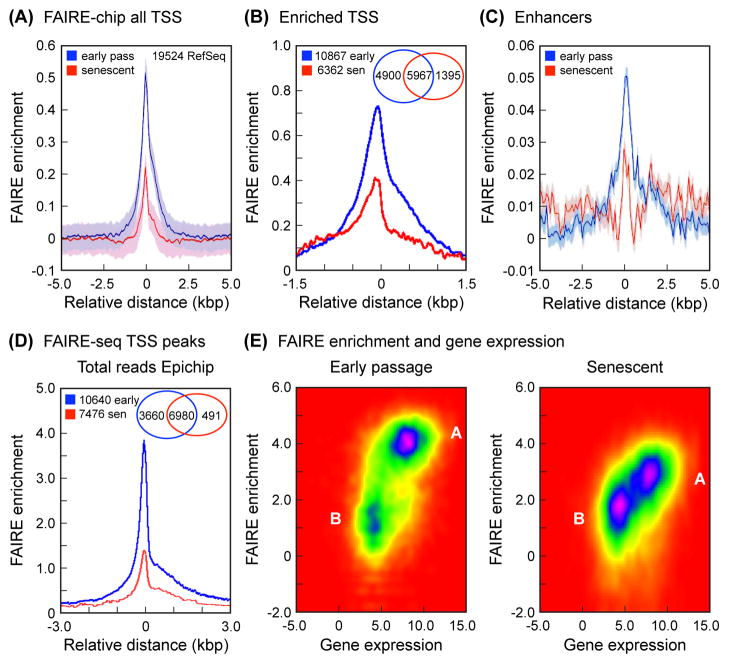
Fig. 1.
FAIRE analysis of TSS and enhancers. (A) FAIRE-chip: average FAIRE signal at TSS decreases in senescent cells. Enrichment (Y-axis) was calculated as the averaged log2 (FAIRE/input) signal for each probe within a 10 kbp window (X-axis) centered on all the selected TSS (corrected for background, which was calculated by interpolating over 530,000 random 5 kbp regions across the genome). The 19,524 RefSeq genes were selected from the 26,083 genes annotated in the hg18 build of the human genome because they contained unique TSS. The interpolated values are shown as a solid line (blue, early passage; red, senescent), and the shaded regions represent the standard deviation envelopes above and below the mean. (B) FAIRE-chip: signal at TSS partitions into 2 clusters and decreases in the enriched cluster in senescent cells. The analysis was performed using 2 clusters and a window of 3 kbp (see Experimental Procedures). Shown here is the FAIRE-enriched cluster, which contains 10,867 genes in early passage cells (blue) and 6,362 genes in senescent cells (red). Interpolated values are shown as in (A). The Venn diagram shows the distribution of genes in the FAIRE enriched cluster: 5,967 genes are enriched in both early passage and senescent cells, 4,900 genes are uniquely enriched in early passage cells, and 1,395 genes are uniquely enriched in senescent cells. (C) FAIRE-chip: enrichment at enhancers decreases in senescent cells. Enrichment was calculated as in (A) using a database of predicted enhancers (Heintzman et al., 2009) (see Experimental Procedures). (D) FAIRE-seq: average FAIRE signal at TSS decreases in senescent cells. All uniquely mapping reads were processed with EpiChIP software (Hebenstreit et al., 2011), using a window of 6 kbp centered on all the TSS in the human genome hg18 or hg19 annotation files. The FDR cutoffs for both signal and noise were set at 0.05. This analysis identified significant TSS peaks in 10,640 genes in early passage cells, and 7,476 genes in senescent cells. The Venn diagram shows the distribution of genes: 6,980 genes have peaks in both early passage and senescent cells, 3,660 genes have peaks only in early passage cells, and 491 genes have peaks only in senescent cells. (E) FAIRE-seq: positive correlation of FAIRE enrichment at TSS with gene expression in early passage (left) and senescent (right) cells. The FAIRE enrichment dataset was intersected with the Affymetrix gene expression dataset and visualized using EpiChIP software. FAIRE enrichment (Y-axis) is shown as reads per gene. Gene expression (X-axis) is shown as PLIER scores. Note that early passage cells contain two well demarcated clusters of genes, one (A) with higher FAIRE and gene expression signals, and one (B) with lower values for both. Cluster A is notably larger. In senescent cells, cluster A loses FAIRE enrichment, and cluster B becomes more prominent.