Fig. 2.
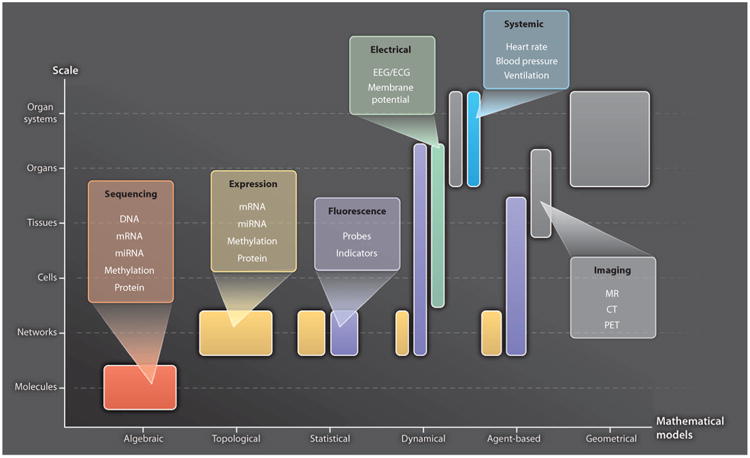
Representative modeling approaches and their data requirements. The abscissa lists different approaches to modeling biological systems at different scales (ordinate). “Algebraic” models describe classes of objects in the genome and their relationships [for example, (107)]. “Topological” models describe molecular wiring diagrams [for example, (108)]. “Statistical” models describe molecular networks as the joint probability distribution of molecular concentrations [for example, (35)]. “Dynamical,” mechanistic models describe the spatiotemporal evolution of biological states using ordinary or partial differential equations [for example, (68)]. “Agent-based” models describe physiological system component interactions using rules, and component location and state evolve in time [for example, (109)]. “Geometrical” models describe anatomic shape [for example, (88)]. Representative data types used in these different modeling approaches and at different biological scales are color-coded. EEG, electroencephalography; ECG, electrocardiography; PET, positron emission tomography.
