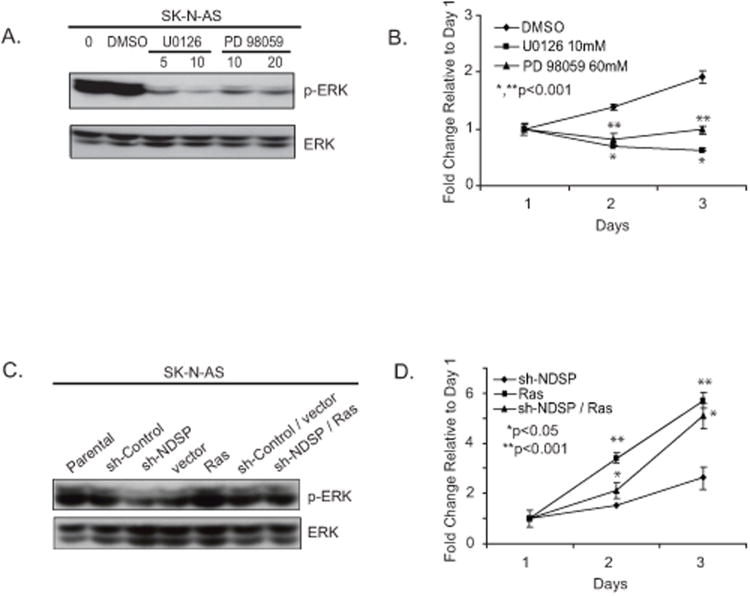
Figure 6.
MEK1/2 inhibitors cause decreased ERK activation in SK-N-AS cells resulting in a prolifereation defect. A, The MEK1/2 inhibitor assays were performed by placing SK-N-AS cells in culture at 3 × 105 cells/well and incubating for 24 hours. Various concentrations of U0126 (5μM, 10μM), PD98059(10μM, 20μM) were added to the cell growth medium and incubation was continued for an additional 24 hours. Dimethyl sulfoxide only was administered as the control treatment. Cells were harvested and immunoblotted for phospho-ERK1/2 and total ERK1/2. B, To test proliferation, SK-N-AS cells were plated in 96-well flat-bottom plates at 1 × 104 cells/well. Twenty-four hours later, various concentrations of U0126 (10μM) or PD98059 (60μM) were added to the cell growth medium and incubation was continued for an additional 3 days. Cytotoxic activity was determined by the CCK-8 assay. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing the U0126 group to the DMSO control. **Indicates statistical significance comparing the PD98059 group to the DMSO control C, In order to test the effects of constitutive ERK1/2 activation, SK-N-AS parental cells were cultured at 3× 105 cells/well and were transiently transfected with sh-control, sh-NDSP, pBabe-vector control (vector), pBabe-RasV12 (Ras), sh-Control/vector, and sh-NDSP/Ras. Forty-eight hours after transfection, the cells were collected and immunoblotted for phospho-ERK1/2 and total ERK1/2. D, The cells were also re-seeded into 96-well plates at 1 × 104 cells/well and allowed to grow for an additional 4 days to compare growth among the experimental groups. These experiments were performed in triplicate and reported as the mean with standard deviations. Student's T-test was used to determine statistical significance with a p-value < 0.05 considered statistically significant. *Indicates statistical significance comparing the sh-NDSP/Ras group to the sh-NDSP group. **Indicates statistical significance between the Ras group and the sh-NDSP group.