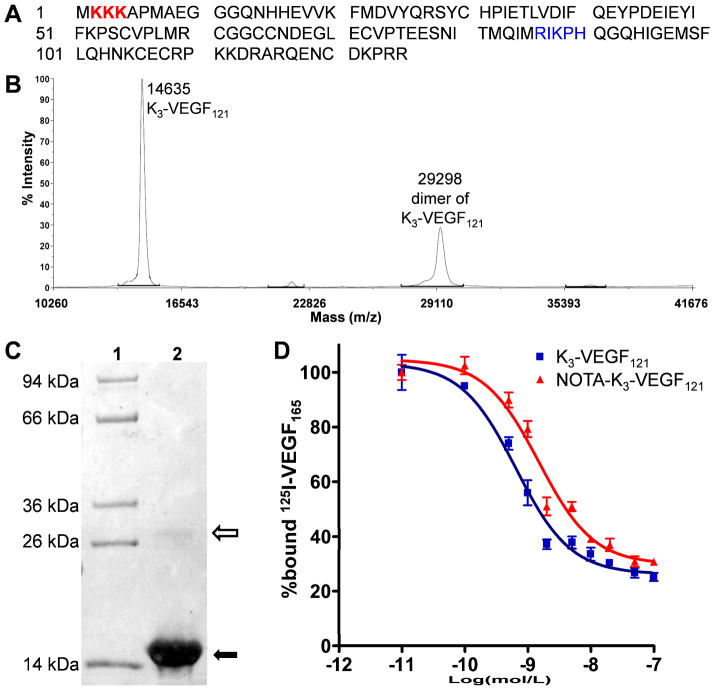
Figure 1.
Characterization of K3-VEGF121 and NOTA-K3-VEGF121. (A) The amino acid sequence of K3-VEGF121. The 3 added lysine residues are coded in red and the amino acid residues involved in VEGFR-2 binding are shown in blue, which contain a lysine residue. (B) Mass spectrometry of K3-VEGF121 which shows the expected peak (14635 Da) as well as the homodimer (29298 Da). (C) SDS-PAGE gel indicated > 95% purity of K3-VEGF121 (solid arrow) with a very light homodimer band (empty arrow). Lane 1: molecular weight marker; Lane 2: K3-VEGF121. (D) Cell binding assay in PAE/KDR cells with 125I-VEGF165 as the competitive ligand. Data represent triplicate samples. The IC50 values are 0.65 and 1.50 nM for K3-VEGF121 and NOTA-K3-VEGF121, respectively.