Figure 5. Kinetics of equilibrium DNA cleavage by K167A and K167C.
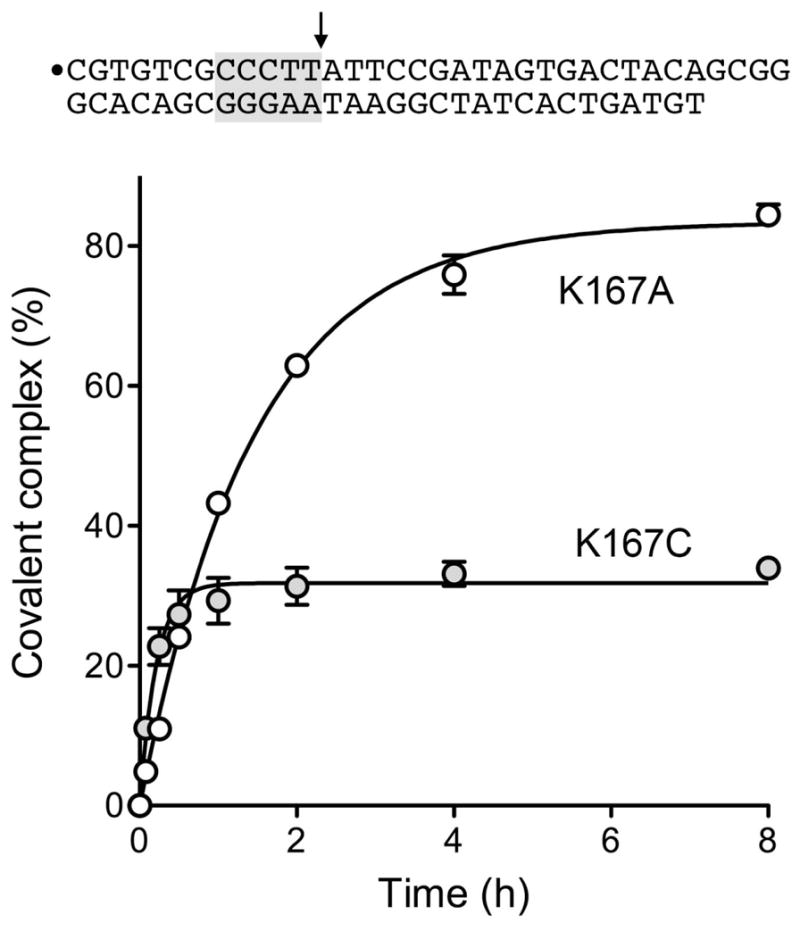
The 34-mer/30-mer equilibrium cleavage substrate is depicted at the top with the 5′ 32P-label denoted by ●, the CCCTT sequence shaded gray, and the cleavage site indicated by the arrow. Reaction mixtures containing (per 20 μl) 50 mM Tris-HCl (pH 7.5), 0.3 pmol of 32P-labeled DNA, and 150 ng (4 pmol) of purified K167A or K167C TopIB were incubated at 37°C. The reactions were initiated by adding TopIB to pre-warmed reaction mixtures. Aliquots (20 μl) were withdrawn at the times specified and the reactions were quenched immediately with SDS. The quenched reactions were treated with 10 μg of proteinase K for 60 min at 37°C, then mixed with an equal volume of 95% formamide/20 mM EDTA, and heat-denatured prior to electrophoresis through a 17% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris-borate, 2.5 mM EDTA). The radiolabeled DNAs and the DNA-peptide adducts derived by proteolysis of the covalent TopIB-DNA complex were quantified by scanning the gel. Covalent adduct formation (expressed as the percent of the total labeled DNA) is plotted as a function of reaction time. Each datum is the average of three separate experiments ±SEM. Non-linear regression curve fits to a pseudo-first order exponential function (executed in Prism) are shown.
