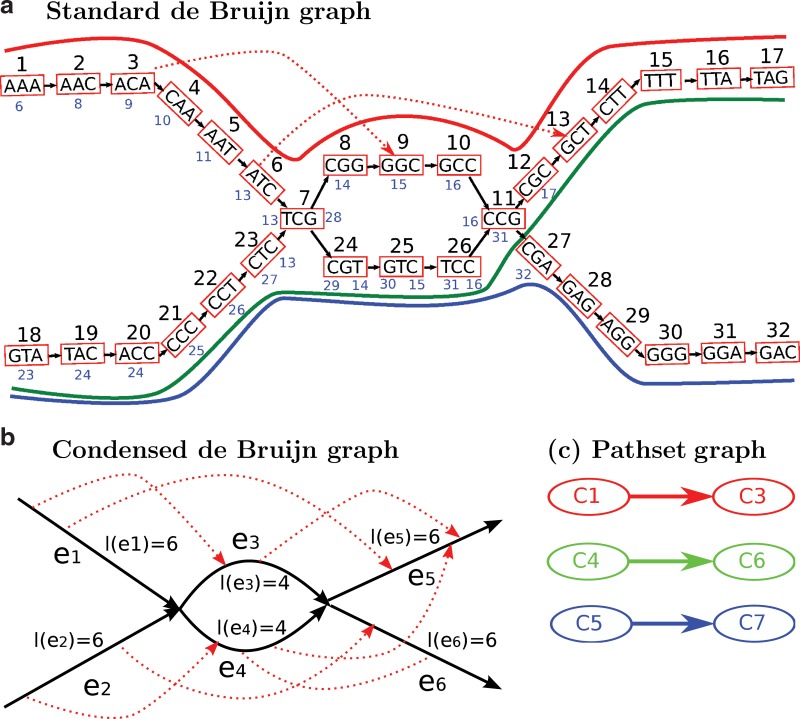
FIG. 1.
From de Bruijn graph to pathset graph. (a) A standard de Bruijn graph and the corresponding mapping of mate-pairs. The number on top of each node is the node ID. The smaller blue numbers below/beside each node are the IDs of the corresponding paired right nodes. The bold red, blue, and green paths show how the genome traverses the graph. (b) The condensed de Bruijn graph with edges corresponding to non-branching paths in the standard de Bruijn graph. The dotted red lines indicate edge-pairs. (c) Pathset graph. Initially there are eight pathsets: C1 = {e1e3e5, e1e4e5}, C2 = {e1e3}, C3 = {e3e5}, C4 = {e2e3e5, e2e4e5}, C5 = {e2e3e6, e2e4e6}, C6 = {e4e5}, C7 = {e4e6}, and C8 = {e2e4}. Using the edge-pair information, we find phantom paths (indicated in boldface) and remove them. After removal of all prefix pathsets (C2 and C8), the pathset graph has six nodes and consists of three edges: C1 → C3 (red path), C4 → C6 (green path), and C5 → C7 (blue path). Each edge in the pathset graph corresponds to a contig; e.g., C1 → C3 spells out the red path (AAACAATCGGCCGCTTTAG).