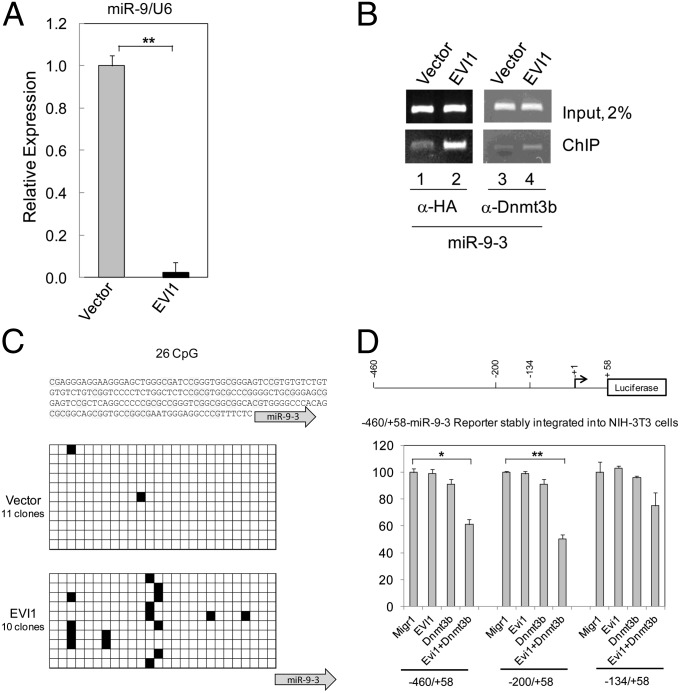
Fig. 4.
EVI1 down-regulates miR-9 in BM cells through DNA methylation. (A) miR-9 expression in BM cells infected with vector or EVI1 and cultured in vitro for 18 d. The expression of miR-9 was determined by qRT-PCR. Expression of miR-9 is taken arbitrarily as 1 for vector-infected BM cells. (B) ChIP assay with 293T cells transiently transfected with vector or EVI1. (C) miR-9-3 promoter methylation analysis in Lin− BM cells infected with vector or EVI1 and cultured in vitro for 10 d. (Top) The analyzed sequence. (Middle and Bottom) There are 26 CpG dinucleotides within the sequenced stretch of DNA, indicated by black (methylated) or empty (unmethylated) cells. (D) EVI1 synergizes with Dnmt3b to repress the regulatory regions of miR-9-3. (Upper) Diagram of a miR-9-3 luciferase reporter gene. Numbers indicate the nucleotide boundaries of the reporter constructs, and they are numbered with respect to the stem-loop start in the miR-9-3 gene taken as +1 and indicated by the arrow. (Lower) Luciferase reporter assay. NIH 3T3 cells stably transfected with reporter plasmids were transiently cotransfected with effector plasmids as shown.