Abstract
Standard whole-genome genotyping technologies are unable to determine haplotypes. Here we describe a method for rapid and cost-effective long-range haplotyping. Genomic DNA is diluted and distributed into multiple aliquots such that each aliquot receives a fraction of a haploid copy. The DNA template in each aliquot is amplified by multiple displacement amplification, converted into barcoded sequencing libraries using Nextera technology, and sequenced in multiplexed pools. To assess the performance of our method, we combined two male genomic DNA samples at equal ratios, resulting in a sample with diploid X chromosomes with known haplotypes. Pools of the multiplexed sequencing libraries were subjected to targeted pull-down of a 1-Mb contiguous region of the X-chromosome Duchenne muscular dystrophy gene. We were able to phase the Duchenne muscular dystrophy region into two contiguous haplotype blocks with a mean length of 494 kb. The haplotypes showed 99% agreement with the consensus base calls made by sequencing the individual DNAs. We subsequently used the strategy to haplotype two human genomes. Standard genomic sequencing to identify all heterozygous SNPs in the sample was combined with dilution-amplification–based sequencing data to resolve the phase of identified heterozygous SNPs. Using this procedure, we were able to phase >95% of the heterozygous SNPs from the diploid sequence data. The N50 for a Yoruba male DNA was 702 kb whereas the N50 for a European female DNA was 358 kb. Therefore, the strategy described here is suitable for haplotyping of a set of targeted regions as well as of the entire genome.
Keywords: genotype, next generation sequencing, phasing
Current whole-genome genotyping technologies such as high-density SNP arrays and whole-genome sequencing can determine a subject’s genotype, but are unable to determine which sequences of interest are located on the same chromosome. The most frequently used method to resolve haplotypes is through inference or statistical computation from parental or population genotypes (reviewed in ref. 1). However, not all haplotypes can be resolved through computational methods, and samples from related individuals may not always be available to the investigator. Several experimental phasing methods have been described. Examples include somatic cell hybrid construction to convert a diploid cell into haploid cell lines (2); large-insert cloning followed by screening of clone pools by either PCR-based genotyping (3) or sequencing (4–6) and isolation of individual chromosomes through chromosome microdissection of lysed metaphase cells on a glass slide (7–9); FACS-mediated single chromosome sorting (10); or the use of a custom microfluidic device (11) followed by amplification of the individual chromosomes and genotyping. The requirement of high-level expertise is a common denominator in these methods, which are not amenable to high-throughput studies or easily transferable to a clinical or diagnostic setting. Alternatively, single-molecule, long-read sequencing technologies such as single molecule real time (SMRT) sequencing, real-time DNA sequencing using fluorescence resonance energy transfer, and nanopore sequencing methods should enable direct phasing of the template DNA (12–14). However, these methodologies are at different stages of development and are either currently still unavailable to investigators or require highly specialized and costly equipment.
Several dilution-based haplotyping methods have also been reported. Single-molecule dilution is followed by amplification and identification of the variants of interest by a variety of methods including restriction fragment length polymorphism analysis, allele-specific PCR, genotyping by mass spectrometry, and Sanger sequencing (15–17). However, none of these methods are able to phase across long distances, and only a limited number of loci can be phased. Furthermore, at single-molecule levels stochastic sampling will result in aliquots receiving zero, one, two, or more copies of the intended target region as dictated by the Poisson distribution (18). Therefore, a significant portion of the aliquots will be uninformative due to either the absence of template or the presence of (greater than) diploid content. One study addressed this by carrying out a PCR prescreen of all aliquots, followed by genotyping of the subset of aliquots containing the region of interest (19). However, a significant number of prescreen PCR reactions are required as each aliquot needs to be cross-referenced against all regions of interest, thereby limiting the number of target regions that can be addressed. A recently published study overcomes these limitations by combining the dilution-amplification method with massively parallel sequencing using Complete Genomics’ service for whole-genome haplotyping (20).
Here we present a method for rapid whole-genome haplotyping. Proof of concept is demonstrated using targeted haplotyping of a 1-Mb contiguous region of the Duchenne muscular dystrophy (DMD) gene. The strategy is subsequently applied to the phasing of two human genomes. The method makes use of single-molecule dilution and takes advantage of the high throughput of next generation sequencing (NGS) platforms. Therefore, the method can be used by any investigator with access to an NGS instrument. In short, genomic DNA is diluted and distributed into multiple aliquots such that each aliquot receives a fraction of one haploid copy; therefore, any position of the genome is likely to be represented by haploid DNA. The DNA template in each aliquot is independently amplified using multiple displacement amplification (MDA) to generate sufficient input material for sequencing library preparation. The amplified products are subsequently converted into barcoded sequencing libraries using Illumina Nextera technology, such that library fragments in each aliquot receive a unique, aliquot-specific barcode, followed either by pooling and massively parallel sequencing or by pooling and targeted enrichment through probe-assisted pull-down and massively parallel sequencing. The association of the same aliquot-specific barcode with adjacent SNPs indicates that the DNA strands carrying these alleles are present within the same aliquot and are therefore highly likely to have been derived from the same starting molecule. The results presented here demonstrate that this strategy can be applied to a set of targeted regions or to an entire genome.
Results
Isolation of Subhaploid DNA Segments.
To assess whether the dilution-amplification method can successfully isolate subhaploid segments of DNA and render these accessible to subsequent manipulation, we compared genotyping data derived from 200 ng of diploid genomic DNA (NA20847, female Gujarati Indian in Houston) to genotyping data from the same genomic DNA sample diluted to an estimated 0.15 haploid copies per aliquot followed by MDA. The genotypes were determined on an Illumina 300K HumanCytoSNP-12 BeadChip (Fig. 1). Scatterplots of the raw A and B intensities were used to indicate the presence of homozygous and heterozygous loci, arbitrarily called A and B, in the starting material. Heterozygous A/B alleles should be present only if more than one haploid copy of that particular region is present in the starting material. Fig. 1 demonstrates the ability of the method to resolve heterozygous SNPs into their haploid components. The scatterplot in Fig. 1A represents raw intensities of the normal diploid individual. Fig. 1B (Left and Right) represents raw intensities of two of the matching dilution-amplification samples. The heterozygous loci in the diploid genomic data, indicated in purple, are concentrated along the x axis and the y axis, indicating that these have been resolved into one of their hemizygous (A/0 or B/0) components by the dilution. Due to the fact that the template DNA is distributed by random sampling, the chance of overlapping fragments of both maternal and paternal descent being present in the same aliquot is subject to Poisson distribution. For a dilution containing 0.15 haploid copies per aliquot and an average minor allele frequency of 0.2 for all SNPs on the 300K HumanCytoSNP-12 BeadChip, the probability of encountering both alleles of a heterozygous SNP within a single aliquot corresponds to 0.72%. The experimental fractions of heterozygous SNP calls in the dilutions shown in Fig. 1B are 1.31 and 0.76%, respectively. Deviations from the theoretical heterozygous fraction are likely due to experimental noise.
Fig. 1.

The dilution method resolves heterozygous SNPs into their haploid components. (A) Raw intensities of an Illumina HumanCytoSNP-12 BeadChip obtained with 200 ng of diploid DNA, demonstrating A/A (red), B/B (blue), and A/B (purple) genotype loci. (B) Raw intensities of two matching dilution-amplification samples at an estimated 0.15 haploid copies per aliquot. The heterozygous loci in the diploid genomic data, indicated in purple, have been resolved into one of their hemizygous (A/0 or B/0) components by the dilution. The percentages of heterozygous SNP calls are indicated.
Accurate, absolute DNA copy-number determination as required for the calculation of the correct dilution is challenging. Furthermore, there exists the possibility that not all template material present in a dilution aliquot is amplified efficiently during the MDA reaction, and therefore less than the original starting quantity will be interrogated. We performed a titration series, read-out on an Illumina 300K HumanCytoSNP-12 BeadChip to assess the effective concentration (Fig. S1A). From the genotyping data, the fraction of SNPs called and the percentage of heterozygous SNPs present in the data can be determined (Fig. S1 B and C). Based on the fraction of SNPs that were detected in this particular experiment, the actionable concentration appears to be lower than the concentration estimated with the intercalating PicoGreen dye. Alternative approaches such as digital PCR (21) or assaying a limited number of dilution aliquots in a short sequencing run or with PCR-based genotyping reactions could be used instead to determine genomic coverage per aliquot. Furthermore, for subsequent experiments inclusion of previously characterized reference samples in PicoGreen measurements was found to be sufficient.
Targeted Analysis of DMD Region.
As a proof-of-principle experiment, we examined a 1-Mb contiguous region of the DMD gene, located on the X chromosome, in two male Coriell genomic DNA samples: NA18507 (22) and HG01377 (23). These samples were sequenced individually after a TruSeq targeted pull-down of the 1-Mb contiguous region of the DMD gene. Because the DMD gene is located on the X chromosome, this allows for direct phasing of the two haplotypes. The two male samples were subsequently combined at equal ratios to create a sample that is artificially diploid for the X chromosome. The DNA mixture was diluted and distributed into 96 aliquots at an estimated 0.2 haploid copies per aliquot. Each aliquot of diluted template DNA was individually amplified with MDA. To confirm hemizygosity and genome coverage of the aliquots, a portion of the amplified material for a subset of aliquots was assayed on an Illumina 300K HumanCytoSNP-12 BeadChip (Fig. S2A). In each aliquot an average 11% of the SNPs assayed were detected of which 1.3% resulted in a heterozygous SNP call. The remainder of the amplified material was converted into barcoded sequencing libraries, pooled, and subjected to targeted pull-down of the 1-Mb contiguous region of the DMD gene. The enriched libraries were subsequently sequenced on an Illumina Genome Analyzer IIx. The DMD region of the undiluted “diploid” DNA mixture was also sequenced to ascertain all heterozygous SNPs within the region.
We first made the heterozygous SNP calls based on the artificial diploid sequencing data. Filtering at Q20, we called 1,265 heterozygous SNPs. At each of these positions we made a consensus call in the data from the two individual male samples to define the haplotypes. In the individual male samples, 55 sites (4.4%) were found to be heterozygous in either one or both of the samples. These heterozygous SNP calls are most likely due to duplications, errors in the reference sequence, or misalignment of off-target reads and were filtered out to yield 1,210 total phased SNPs spanning 980 kb.
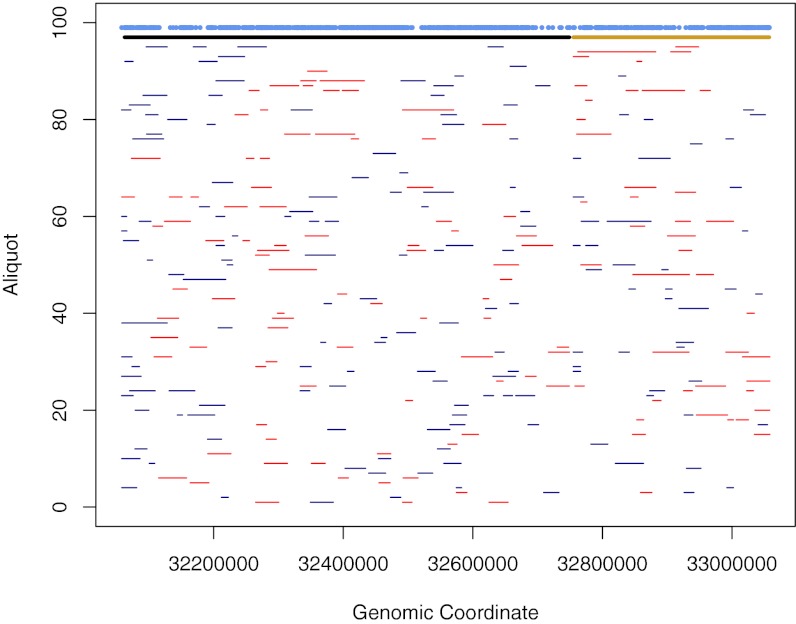
We proceeded to extract out the consensus calls within the haploid data from the dilution experiments and merged these into haplotype blocks (for further details, see SI Text, section S1). This produced two haplotype blocks with lengths 303 and 687 kb, and 1,200 of the 1,210 SNPs phased into either one of these blocks (Fig. 2). Genomic context of the 6-kb gap separating the two blocks negatively affects alignment in both the diploid sequencing data and the dilution data as well as in standard whole-genome sequencing data as the region contains a large stretch of repetitive sequence (Fig. S2D). Furthermore, the gap contains no polymorphic sites in the diploid sequence data (Fig. 2), impeding phasing of this stretch. Comparing the phased haplotypes to the known haplotypes from the sequencing data of the two individual samples, we estimate that, for the longer block, the phasing accuracy was 99% (803 of 812 SNPs correctly phased, no switch errors detected) and, for the shorter block, the phasing accuracy was 97% (375 of 388 sites correctly phased, no switch errors detected).
Fig. 2.
Illustration of the DMD haplotype blocks. Contiguous segments detected in the individual dilution aliquots are indicated at the bottom part of the panel. Bars in red correspond to the HG01377 haplotype, and bars in blue correspond to the NA18507 haplotype. The two merged haplotype blocks are indicated with the black and gold bars at the top. Blue overlapping circles across the top indicate heterozygous SNP positions in the diploid sequencing data.
Whole-Genome Haplotyping.
Because experiments with our DMD system showed feasibility of the approach, we applied the method to phasing of an entire human genome [NA18506, male Yoruba in Ibadan, Nigeria (YRI)] with a few protocol modifications (SI Text, section S3 and Table S1). Template DNA input was increased to 0.4 haploid copies, based on modeling experiments carried out with the DMD data (SI Text, section S2). In an initial experiment, we prepared two independent collections of 96 dilution aliquots each. The combined sequencing data of all 192 aliquots yielded 81.6-fold average genome coverage and covered 90.3% of the 3.1-Gb human genome (Table S2). Of the 3,093,078 heterozygous SNPs, 95.6% were phased into a total of 9,243 blocks (Table 1). The average haplotype block is 264 kb, with a maximum of 4.8 Mb and an N50 of 702 kb. Compared with haplotypes derived from diploid whole-genome sequencing of the family trio (father, mother, and son), phase could be verified for 80% of the SNPs. For the remaining 20%, trio-based phasing was unable to determine phase. These SNPs were subsequently excluded from evaluation. The average block accuracy when disregarding switch errors was determined to be 85%. However, the overall accuracy when considering switch errors was 99.6% with a switch error rate of 0.74%. A more detailed explanation of accuracy calculations can be found in SI Text, section S4 and Table S3.
Table 1.
Summary of YRI and CEPH whole-genome phasing haplotype block metrics and accuracy
| Ethnicity (mean genomic depth) | No. aliquots | No. SNPs phased | No. SNPs used for phasing | % SNPs phased | Total no. blocks | Average haplotype block size, kb | Longest haplotype block size, Mb | N50, kb | Average block accuracy, %* | Overall accuracy considering switch errors, %* | Switch error rate, %* |
| YRI (82×) | 192 | 2,958,022 | 3,093,078 | 95.6 | 9,243 | 264 | 4.8 | 702 | 85 | 99.60 | 0.74 |
| CEPH (58×) | 192 | 2,102,215 | 2,164,688 | 97.1 | 14,880 | 141 | 2.5 | 358 | 90 | 99.65 | 0.61 |
| CEPH† (115×) | 192 | 2,132,069 | 2,164,688 | 98.5 | 10,503 | 221 | 3.1 | 542 | 88 | 99.67 | 0.61 |
| CEPH‡ (58×) | 192 | 2,002,115 | 2,095,856 | 95.5 | 15,132 | 139 | 3.0 | 348 | 87 | 99.74 | 0.55 |
| CEPH¶ (29×) | 96 | 2,038,619 | 2,164,688 | 94.2 | 22,919 | 85 | 1.7 | 197 | 90 | 99.57 | 0.78 |
*For further explanation of accuracy calculations, see SI Text, section S4 and Table S3.
†Increased depth (two flow cells combined).
‡Dilution data only.
¶Down-sampled to 96 aliquots.
The protocol as optimized with the Yoruban genome was subsequently applied to a genome of European descent (NA12878, female Utah resident with ancestry from northern and western Europe, CEPH/UTAH). A total of 192 aliquots at 0.4 haploid copies of genomic DNA each were used and sequenced in duplicate on two flow cells on a HiSeq 2000 (flow cells FC-A and FC-B). FC-A yielded 206 Gb total data and 179 Gb of mapped data (i.e., 87% aligned) for an average genomic coverage of 58× and detection of 90.3% of the 3.1-Gb human genome (Table S2). Combining the data of both flow cells gave 115× depth but did not significantly increase the fraction of the genome detected. Individual aliquots yielded on average 900 Mb worth of sequence, derived from 8.4% or 260 Mb of coverage of the human genome at a mean depth of 3.5×. The lower-than-anticipated haploid content per aliquot could be due to inefficient amplification of the DNA in each aliquot, tagmentation efficiency and its dependency on double-stranded DNA [MDA generates both single-stranded and double-stranded DNA (24)], and/or overestimation of the DNA concentration. Of the 260-Mb genomic equivalents, 226 Mb was assigned to contiguous segments by the targetcut function of the SAMtools package. The remaining 34 Mb are isolated reads at low depth and are therefore not assigned to contiguous segments. A distribution of the haploid content in each dilution aliquot as estimated by the sum of all targetcut segments per dilution can be found in Fig. S3A. A histogram of the targetcut segment size distribution, reflective of the size of the genomic DNA template molecules, is shown in Fig. S3B. The average targetcut segment size is 13.8 kb.
In the matching 30× diploid sequencing data, 2,164,688 heterozygous SNP calls were made. Of these heterozygous SNPs, 97.1 or 98.5% were phased in the 58× or 115× data, respectively (Table 1). On average, each SNP position was detected in 12.4 independent dilution aliquots (Fig. S3C), and the mean depth per detected SNP per aliquot was 4.1× (Fig. S3D). Phasing of the data generated with a single flow cell resulted in a total of 14,880 haplotype blocks with an average length of 141 kb, a maximum length of 2.5 Mb, and an N50 of 358 kb. The decreased average block size and increased number of blocks compared with the data generated with NA18506 reflects the fact that the NA12878 genome exhibits lower levels of heterozygosity. A snapshot of the individual contiguous segments found in the 192 aliquots and the merged haplotype blocks for a section on chromosome 22 is shown in Fig. 3. The gaps in between the merged haplotype blocks coincide with decreased heterozygous SNP density as indicated by the blue overlapping circles. Therefore, even though some contiguous segments span the areas underlying the gaps, a lack of heterozygous SNPs in the overlapping sections prohibits the merging of these segments, causing a break in the haplotype block. When the sequencing depth was doubled by combining the data of FC-A and FC-B, the total number of blocks decreased to 10,503. The mean block size increased to 221 kb, the maximum length to 3.1 Mb, and the N50 to 542 kb.
Fig. 3.
An example of the individual targetcut fragments found in the 192 aliquots and the merged Refhap haplotype blocks for a portion of chromosome 22 in the CEPH data. Red bars represent the contiguous segments detected in the individual dilution aliquots, indicated on the y axis. The black bars at the top indicate the merged haplotype blocks. Blue overlapping circles across the top indicate heterozygous SNP positions in the diploid sequencing data and emphasize the lack of heterozygosity in the gap areas.
The phasing results were compared with family trio-based phasing, which allowed for evaluation of 79% of the phased SNPs. Average block accuracy, when not considering switch errors of the data generated with FC-A alone, was determined to be 90% (Table 1), whereas that of the combined data of FC-A and FC-B corresponded to 88%. However, when considering switch errors, the overall accuracy was determined to be 99.65 and 99.67% with switch error rates of 0.61 and 0.61% for the 58× and 115× data, respectively. Therefore, whereas increasing sequencing depth increased the haplotype block size, the accuracy of the phasing was largely unaffected.
When the compiled dilution data obtained with FC-A was used to make the heterozygous SNP calls instead of the matching 30× standard diploid sequencing data, 2,095,856 heterozygous SNP calls, or 96.8% of the heterozygous SNPs detected in the standard diploid sequencing data, could be made (Table 1). Of these heterozygous SNPs, 2,002,115 were successfully phased for a phasing rate of 95.5%. The total number of haplotype blocks was 15,132 with a mean size of 139 kb, a maximum size of 3.0 Mb, and an N50 of 348 kb. Whereas the overall accuracy considering switch errors and the switch error rate (99.74 and 0.55%, respectively) is similar to the accuracy obtained with the combination of standard diploid sequencing data and FC-A dilution data (99.65% and 0.61% respectively), the average size of the haplotype blocks is decreased. Furthermore, because not all heterozygous SNPs have been detected, certain errors may be masked.
To examine the number of dilution aliquots required at 0.4 haploid copies each, we randomly selected data of a subset of 96 aliquots from the original 192 indices. This corresponds to fewer indices and decreased overall sequencing depth. Modeling experiments resulted in phasing of 94.2% of the heterozygous SNPs, 22,919 haplotype blocks with a mean haplotype block size of 85 kb, 99.57% overall accuracy, and a switch error rate of 0.78%, indicating that more dilution aliquots are required to achieve longer, more accurate haplotype blocks (Table 1).
Of the 36,214 genes, pseudogenes, and ORFs that are encoded in the human genome, we were able to fully phase 27,674 or 76% (Table 2). This included both exonic and intronic regions. A total of 20,460 of those genes contain either zero or one heterozygous site and are therefore considered to be effectively phased. Approximately 15% of genes were partially phased, defined as having greater than 70% of the heterozygous SNPs phased and greater than 70% of the bases composing the gene in the largest haplotype block. The remaining 9% were determined to remain unphased as either less than 70% of the heterozygous SNPs were phased or less than 70% of the bases composing the gene were in the largest haplotype block. A region of great interest with regards to haplotyping is the HLA region on chromosome 6. Of the 12,831 heterozygous SNPs found in the entire 4.8-Mb region, 12,003 were successfully phased into eight haplotype blocks with a mean block size of 606 kb, covering 99.4% of all bases. The largest haplotype block spanned 1.8 Mb. Of the genes within the region, 79% were fully phased, 19% were partially phased, and 2% remained unphased (Table 3). Of the actual HLA genes, 52% were completely phased, 40% partially phased, and 8% unphased. Further details of each individual HLA gene are shown in Table S4.
Table 2.
Genome-wide genic phasing results
| Phasing classification | Genes, n (%) |
| Total genes | 36,214 (100) |
| Effectively phased genes* | 20,460 (56.5) |
| Fully phased genes | 7,214 (19.9) |
| All heterozygous SNPs phased, entire region covered by one haplotype block | 6,122 (16.9) |
| All heterozygous SNPs phased, less than entire region covered by one haplotype block | 1,092 (3.0) |
| Partially phased genes | 5,420 (15.0) |
| More than 70% of heterozygous SNPs phased, entire region covered by one haplotype block | 4,214 (11.6) |
| More than 70% of heterozygous SNPs phased, more than 70% of bases in largest haplotype block | 1,206 (3.3) |
| Unphased genes | 3,120 (8.6) |
| Less than 70% heterozygous SNPs phased | 1,265 (3.5) |
| More than 70% heterozygous SNPs phased, less than 70% of bases in largest haplotype block | 1,855 (5.1) |
*Genes containing zero or one heterozygous SNP.
Table 3.
Phasing results for genes in the 4.8-Mb HLA region on chromosome 6
| Phasing classification | All genes in HLA region, n (%) | HLA genes, n (%) |
| Total genes | 216 (100) | 25 (100) |
| Effectively phased genes* | 71 (32.8) | 1 (4.0) |
| Fully phased genes | 99 (45.8) | 12 (48.0) |
| All heterozygous SNPs phased, entire region covered by one haplotype block | 98 (45.4) | 12 (48.0) |
| All heterozygous SNPs phased, less than entire region covered by one haplotype block | 1 (0.4) | 0 (0) |
| Partially phased genes | 42 (19.4) | 10 (40.0) |
| More than 70% of heterozygous SNPs phased, entire region covered by one haplotype block | 39 (18.0) | 10 (40.0) |
| More than 70% of heterozygous SNPs phased, more than 70% of bases in largest haplotype block | 3 (1.4) | 0 (0) |
| Unphased genes | 4 (1.8) | 2 (8.0) |
| Less than 70% heterozygous SNPs phased | 3 (1.4) | 2 (8.0) |
| More than 70% heterozygous SNPs phased, less than 70% of bases in largest haplotype block | 1 (0.4) | 0 (0) |
*Genes containing zero or one heterozygous SNP.
Discussion
We have shown here that genomic DNA at subhaploid quantities can be rendered accessible to sequencing library preparation through amplification using an MDA approach in standard laboratory equipment and that the association of aliquot-specific barcodes subsequently allows for haplotyping of large genomic regions. Furthermore, the method is platform agnostic as any type of standard downstream sequencing library preparation can be applied. It is therefore a method accessible to any researcher. The added cost associated with obtaining phase in addition to whole-genome sequencing is largely determined by the extra sequencing required and will decrease as sequencing costs continue to decrease; as library preparation methods evolve and are miniaturized, allowing for smaller reaction volumes; and as improvements to amplification methods of subhaploid quantities lead to a reduction in overamplification bias. Examples of the latter include adaptor-mediated universal long-range PCR methods or multiple annealing and looping-based amplification cycles (25).
The bias introduced during the MDA reaction remains an important challenge. It is well known that MDA randomly overreplicates loci (26, 27). In a diploid context, a side effect of the overamplification is allelic dropout (ADO) as a locus on one chromosome may be overamplified relative to the same locus on the other chromosome, leading to overrepresentation of one allele in the data. Furthermore, the deviation from the expected 50:50 ratio makes heterozygous SNP calling challenging. In the whole-genome haplotyping application of MDA, however, the starting material is subhaploid and is sampled in multiple independent amplification reactions. Due to the random nature of the overamplification, there is no correlation between the read depths of the same region between independent amplification reactions. Therefore, increasing the number of aliquots smooths out the average read depth across the combined data and reduces ADO. This results in an improved heterozygous SNP calling rate of the stand-alone dilution data. Indeed, the compiled CEPH whole-genome dilution haplotyping data allowed for the detection of 97% of all heterozygous SNPs identified in the matching standard 30× diploid sequencing data. We were subsequently able to phase the genome with only a relatively small reduction in haplotype block sizes compared with the data paired with the standard 30× diploid sequencing data. To make heterozygous SNP calls from the dilution data, each SNP has to be present in a minimum of two aliquots. Therefore, SNPs detected with a lower aliquot frequency are automatically filtered out, subsequently improving the confidence of the homozygous calls. Increasing the number of dilutions should improve the data further and potentially allows for whole-genome haplotyping without matching 30× diploid sequencing data.
We have determined that the optimal dilution level that allows for accurate haplotyping, while simultaneously keeping the number of dilution aliquots required at a reasonable scale, is 0.2–0.4 haploid copies of a human genome per aliquot (SI Text, section S5 and Table S5). Increasing total sequencing depth was shown to increase haplotype block size, but did not significantly improve the accuracy of the phasing results. The biggest impact came from increasing the number of dilution aliquots. Doubling the number of dilution aliquots from 96 to 192 increased the average haplotype block size 3.7- or 1.6-fold for the YRI and CEPH genomes, respectively, and decreased the number of haplotype blocks 3.4- or 1.5-fold, respectively (Table S1 and Table 1). These haplotyping metrics can surely be improved upon by increasing the number of dilution aliquots or by using higher-molecular-weight DNA template molecules that are more likely to span SNP deserts (20) (SI Text, section S6 and Table S6). Furthermore, most of the data analysis was performed with publicly available algorithms that were not custom-designed for these experiments. Optimization of algorithms will likely yield improvements in phasing accuracy and haplotype block length. As protocols for whole-genome haplotyping mature, analysis pipelines are expected to evolve along with method development.
We have shown whole-genome haplotyping to be successful using the method presented here. Average haplotype block sizes ranged from 140 kb (CEPH) to 264 kb (YRI), depending on the level of heterozygosity of the genome assayed. Human genes span, on average, 20–50 kb in the genome although examples of greater than 100 kb have been identified. Therefore, the method described above is highly likely to be successful in phasing intragenic heterozygous SNPs. The majority of phasing interest lies in accurately phasing all heterozygous sites within a single gene and its regulatory sequences because these will all affect the same transcript and protein molecule. Closer inspection of the genic regions showed that only 9% of genes remained unphased whereas the vast majority was either fully or partially phased. The method that we have presented here is therefore applicable to both haplotyping of targeted regions and whole-genome haplotyping.
Materials and Methods
Sample.
Genomic DNA samples (NA20847, NA18507, HG01377, NA18506, and NA12878) were obtained from Coriell Cell Repositories. Genomic DNA concentrations were measured with Quant-iT PicoGreen dsDNA Reagent (Life Technologies). For the DMD haplotyping experiment, equal quantities of NA18507 and HG01377 were combined.
Multiple Displacement Amplification.
For microarray and DMD proof-of-concept experiments, DNA was diluted to the specified number of haploid copies per 3 µL of water and distributed into multiple wells of a 96-well microtiter plate. To each well, 3 µL of Buffer D2 (Qiagen) was added and incubated for 10 min at 4 °C. Three microliters of Stop Solution was added, followed by the addition of 33 µL of Mastermix containing 30 µL of Reaction Buffer, 2 µL DNA polymerase, and 1 µL of a 7.5-mM 9-mer pool containing 6,000 oligonucleotides designed specifically for the human genome (Illumina). The reactions were incubated for 90 min at 30 °C and heat-inactivated for 3 min at 65 °C. The MDA products were purified using ZR-96 DNA Clean and Concentrator-5 plates (Zymo Research) according to the manufacturer’s protocol and eluted in 12 µL of water.
For whole-genome haplotyping, genomic DNA was diluted to 0.4 haploid copies per 1 µL in water. A total of 1 µL was distributed into the wells of 96-well microtiter plates. DNA was denatured by addition of 1 µL of Buffer D1 (Qiagen) and incubation for 3 min at room temperature. The reaction was neutralized by the addition of 2 µL of Buffer N1. Aliquots subsequently received 16 µL of Mastermix (Illumina) containing either DNA hexamers (Fermentas) or RNA hexamers (Integrated DNA Technologies) and 1 µL DNA polymerase. The reactions were incubated for 90 min at 30 °C and heat-inactivated for 3 min at 65 °C. Single-stranded DNA was removed by treatment with S1 nuclease (Fermentas) according to the manufacturer’s instructions. The final MDA products were purified using ZR-96 DNA Clean and Concentrator-5 plates (Zymo Research) and eluted in 20 µL of water.
Infinium Genotyping Assays.
Infinium genotyping assays using 300K HumanCytoSNP-12 BeadChips (Illumina) were run using 4 µL of each purified MDA product according to manufacturer’s instructions. BeadChips were scanned on an iScan, and data were analyzed with GenomeStudio (Illumina).
Sequencing Library Preparation.
For the DMD experiment, 10 µL of MDA product or 50 ng of genomic DNA was converted with Nextera v1 technology according to the manufacturer’s protocol (EpiCentre Biotechnologies). Sequencing libraries of the whole-genome haplotyping MDA products were generated using Nextera v2 technology according to the manufacturer’s protocol (Illumina). Aliquot-specific barcodes were added during the PCR. Library pools were purified with AMPureXP beads at a 0.6 ratio according to the manufacturer’s guidelines (Beckman Coulter Genomics).
Targeted Enrichment.
A probe pool was designed for the targeted pull-down of a 1-Mb contiguous region of the DMD gene. The biotinylated probes were 80 nt long and designed to hybridize to the 5′ region of the DMD gene at 190- to 370-bp intervals. After pooling, the sequencing libraries were enriched for the 1-Mb DMD gene region following the protocol of the TruSeq Custom Enrichment Kit (Illumina).
Next Generation Sequencing.
Enriched, indexed libraries for the DMD proof-of-concept experiment were sequenced on a Genome Analyzer IIx using 75-cycle paired-end sequencing (Illumina). Each lane contained one pool of 12 indexed MDA samples. Indexed whole-genome haplotyping libraries were sequenced on a HiSeq2000 using 101-cycle paired-end, dual-indexing sequencing.
Data Analysis.
The raw sequencing data were converted to fastq format and aligned to a human reference genome (hg19) using Casava1.8 with default parameters (www.illumina.com/documents/products/technotes/technote_eland_variantcalling_improvements.pdf). Variant calling was performed on the 30× sequencing data of each of the undiluted samples NA18507, HG01377, NA18506, and NA12878 as well as the artificial diploid sample of the DMD experiment using Casava1.8. Filtering at SNP quality score 20, heterozygous SNPs detected in the (artificial) diploid sample were used to create a reference set of phase-informative SNPs for haplotyping of the dilution samples.
To define the haplotype contained within each dilution sample, we identified individual stretches of DNA using the targetcut function of SAMtools (version 0.1.18), which detects regions of contiguous coverage. At all of the previously identified heterozygous positions, we made a consensus base call using a simple heuristic approach. A base is called “homozygous” if either of the following conditions is true: (i) only one of the two alleles is observed or (ii) both alleles are observed but the more commonly called base is observed with at least a five-times-higher frequency. A base is called heterozygous if both alleles are observed at least twice and the more commonly observed allele occurs less than five times as often as the less commonly observed allele. The DNA segments were subsequently combined to create long-range haplotypes. First, to remove regions with overlapping haplotypes from the dilution data, we broke down the blocks defined above to include only segments of stretches of homozygous calls that start at either end of the identified fragments. For example, 20 consecutive calls where the fifth site is heterozygous and the remaining sites are homozygous will be broken into two segments where one contains the first four consensus calls and the other contains the 6th through 20th calls. If a segment was found to contain more than one heterozygous SNP call, the area in between the heterozygous SNP calls was excluded from further analysis. This serves to remove potential switch errors due to overlapping haplotypes in the diluted DNA. These filtered haplotype blocks were then combined into long-range haplotypes using RefHap (6).
Supplementary Material
Acknowledgments
We thank Thomas Royce for insightful discussions, Tobias Mann for DMD pull-down probe design, and Luana McAuliffe and Samantha Cooper for bioinformatics support.
Footnotes
Conflict of interest statement: All authors are current employees and shareholders of Illumina, Inc.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) (accession no. SRP018998).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218696110/-/DCSupplemental.
References
- 1.Browning SR, Browning BL. Haplotype phasing: Existing methods and new developments. Nat Rev Genet. 2011;12(10):703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas JA, Boehnke M, Gillanders E, Trent JM, Gruber SB. Experimentally-derived haplotypes substantially increase the efficiency of linkage disequilibrium studies. Nat Genet. 2001;28(4):361–364. doi: 10.1038/ng582. [DOI] [PubMed] [Google Scholar]
- 3.Burgtorf C, et al. Clone-based systematic haplotyping (CSH): A procedure for physical haplotyping of whole genomes. Genome Res. 2003;13(12):2717–2724. doi: 10.1101/gr.1442303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29(1):59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suk E-K, et al. A comprehensively molecular haplotype-resolved genome of a European individual. Genome Res. 2011;21(10):1672–1685. doi: 10.1101/gr.125047.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duitama J, et al. Fosmid-based whole genome haplotyping of a HapMap trio child: Evaluation of single individual haplotyping techniques. Nucleic Acids Res. 2012;40(5):2041–2053. doi: 10.1093/nar/gkr1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Höckner M, Erdel M, Spreiz A, Utermann G, Kotzot D. Whole genome amplification from microdissected chromosomes. Cytogenet Genome Res. 2009;125(2):98–102. doi: 10.1159/000227832. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, et al. Direct determination of molecular haplotypes by chromosome microdissection. Nat Methods. 2010;7(4):299–301. doi: 10.1038/nmeth.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitada K, Taima A, Ogasawara K, Metsugi S, Aikawa S. Chromosome-specific segmentation revealed by structural analysis of individually isolated chromosomes. Genes Chromosomes Cancer. 2011;50(4):217–227. doi: 10.1002/gcc.20847. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Chen X, Wong WH. Completely phased genome sequencing through chromosome sorting. Proc Natl Acad Sci USA. 2011;108(1):12–17. doi: 10.1073/pnas.1016725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29(1):51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 13.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 14.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19(R2):R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 15.Ruano G, Kidd KK, Stephens JC. Haplotype of multiple polymorphisms resolved by enzymatic amplification of single DNA molecules. Proc Natl Acad Sci USA. 1990;87(16):6296–6300. doi: 10.1073/pnas.87.16.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul P, Apgar J. Single-molecule dilution and multiple displacement amplification for molecular haplotyping. Biotechniques. 2005;38(4):553–554, 556, 558–559. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- 17.Ding C, Cantor CR. Direct molecular haplotyping of long-range genomic DNA with M1-PCR. Proc Natl Acad Sci USA. 2003;100(13):7449–7453. doi: 10.1073/pnas.1232475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens JC, Rogers J, Ruano G. Theoretical underpinning of the single-molecule-dilution (SMD) method of direct haplotype resolution. Am J Hum Genet. 1990;46(6):1149–1155. [PMC free article] [PubMed] [Google Scholar]
- 19.Konfortov BA, Bankier AT, Dear PH. An efficient method for multi-locus molecular haplotyping. Nucleic Acids Res. 2007;35(1):e6. doi: 10.1093/nar/gkl742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487(7406):190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24(6):680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 25.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99(8):5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3(9):1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.