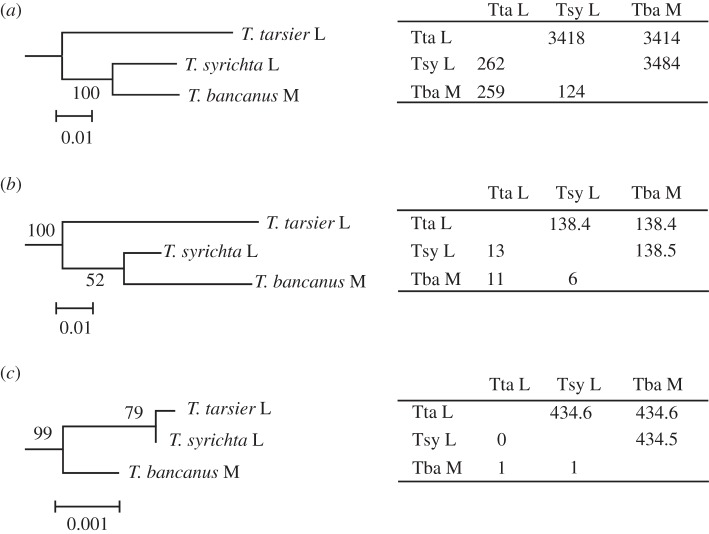
Figure 2.
Phylogenetic trees of the tarsier L/M opsin gene using (a) introns 3 and 4, (b) the synonymous sites of exons 3, 4 and 5, and (c) the non-synonymous sites of exons 3, 4 and 5. For each species by species comparison, the length of the nucleotide sequence (bp) used and the number of nucleotides differing between each pair are presented, above and below the diagonal, respectively, in the tables adjacent to each phylogeny. The bootstrap probability is given at each node. We determined the phylogenetic root for tree (a) using the common marmoset L/M opsin gene (Ensembl Marmoset release 70, Genome assembly C_jacchus3.2.1 (GCA 000004665.1) (http://www.ensembl.org/Callithrix_jacchus/Info/Index), X:140703132–140704540 for intron 3 and X:140704707–140706490 for intron 4) and for trees (b) and (c) using three spectral alleles of the common marmoset L/M opsin gene (GenBank AB046546–AB046548) as the out-group for each tree. Scale bars indicate the number of nucleotide substitutions per site.