Figure 2.
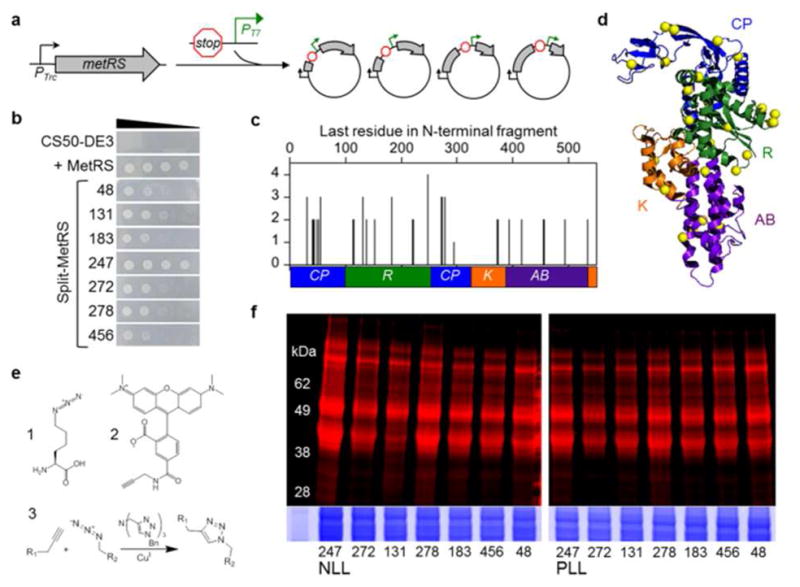
Identifying fragmented MetRS variants that metabolically label proteins with Anl. (a) DNA that contains a terminator, promoter (PT7), and ribosomal binding site was randomly inserted into the MetRS gene to create a library of vectors that express two-piece MetRS variants8. (b) Growth of E. coli CS50-DE3 cells in the absence of Met is compared for cells that express a functional MetRS truncation comprised of residues 1-548 and split MetRS variants. Ten-fold serial dilutions of cells were spotted on M9-agar plates and growth was imaged after 24 h. (c) Complementation strengths of fragmented MetRS variants, which were named based on the last residue encoded by their N-terminal fragments. (d) Color coding of the connective polypeptide [CP, blue], Rossmann fold [R, green], KMSKS [K, orange], and anticodon-binding [AB, purple] domains maps the locations of backbone fission (yellow spheres) in functional variants onto the MetRS structure11. (e) Structure of Anl (1), which can be detected in proteins after reaction with alkyne-functionalized tetramethylrhodamine (TAMRA) dye (2). Copper-catalyzed azide-alkyne cycloaddition (3) results in a stable triazole linkage. (f) Cells expressing the split NLL-MetRS and PLL-MetRS from the vectors selected using bacterial complementation were grown in the presence of Anl, and cell lysates were treated with TAMRA-alkyne dye. In-gel fluorescence image shows TAMRA labeling, which indicates Anl incorporation into cellular proteins. Colloidal blue staining (bottom) was performed to compare protein loading across lanes.
