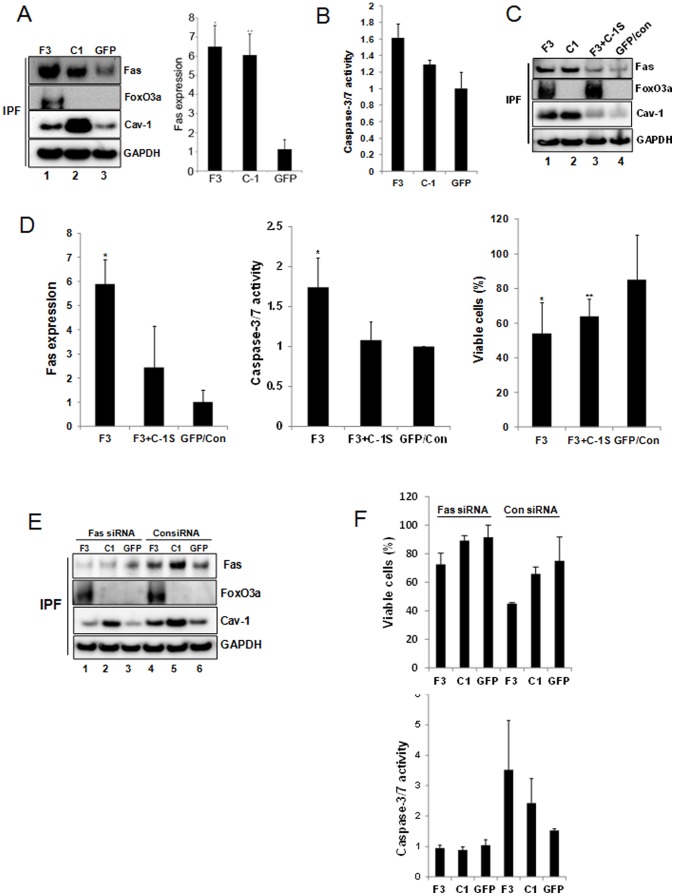
Figure 6. Forced expression of FoxO3a increases Fas expression via cav-1 in IPF fibroblasts and promotes apoptosis on polymerized collagen matrix.
A. IPF fibroblasts infected with adenovirus over-expressing FoxO3a (F3), cav-1 (C-1) or empty vector GFP were cultured on polymerized collagen for 48 h in serum free medium. Left panel, Fas, FoxO3a, cav-1 and GAPDH expression were measured by Western blot analysis. Three independent assays are represented. Right panel. Fas/GAPDH protein expression ratio was quantified. *p = 0.02 versus GFP. **p = 0.05 versus GFP. B. IPF fibroblasts infected with adenovirus over-expressing FoxO3a (F3), cav-1 (C-1) or empty vector GFP were cultured on polymerized collagen for 48 h and caspase-3/7 activity was measured. Results were obtained from triplicates. C. IPF fibroblasts infected with adenovirus over-expressing FoxO3a (F3), cav-1 (C1), FoxO3a in the presence of 10 nM of Cav-1 siRNA (F3+C-1S), or empty vector GFP with 10 nM control siRNA (GFP/Con) were cultured on polymerized collagen for 48 hrs. Fas, FoxO3a, cav-1 and GAPDH expression were examined by Western blot analysis. Blot represents 3 independent assays. D. IPF fibroblasts infected with adenovirus expressing FoxO3a alone (F3) or IPF fibroblasts over-expressing FoxO3a in the presence of 10 nM of Cav-1 siRNA (F3+C-1S) or GFP/Control siRNA (GFP/Con) were cultured on polymerized collagen for 48 hrs. Left panel, Fas expression was quantified. *p = 0.03 versus GFP/Con. Results were obtained from 3 independent assays. Middle panel, Caspase-3/7 activity was measured. *p = 0.01 versus GFP/Con. Results were obtained from triplicates. Right panel, viable cells were quantified. *p = 0.01, **p = 0.05 versus GFP/Con, respectively. Results were obtained from quadruplicates. E–F. 2×105 IPF fibroblasts infected with adenovirus over-expressing FoxO3a (F3), cav-1 (C1) or empty vector GFP were cultured on polymerized collagen in the presence of 10 nM Fas or control siRNA for 48 h in serum free medium. Fas, FoxO3a, cav-1 and GAPDH expression were measured by Western blot analysis (E). Cell viability (F, upper panel) and caspase-3/7 activity (F, lower panel) were measured as described in Materials and Methods. Results were obtained from triplicates.