RhoGEF12 is required for stretch-induced RhoA activation, and its absence protects mice against overload-induced heart failure.
Abstract
Structural cardiac remodeling, including hypertrophy and fibrosis, plays a crucial role in the pathogenesis of heart failure. In vitro studies suggested a role of the small GTPase RhoA in hypertrophic cardiomyocyte growth, but neither the molecular mechanisms leading to RhoA activation nor their relevance in vivo are known. We use here a mass spectrometric approach to identify Rho guanine nucleotide exchange factors (RhoGEFs) activated during cardiac pressure overload in vivo and show that RhoGEF12 is a central player during cardiac remodeling. We show that RhoGEF12 is required for stretch-induced RhoA activation and hypertrophic gene transcription in vitro and that its activation depends on integrin β1 and heterotrimeric G proteins of the G12/13 family. In vivo, cardiomyocyte-specific deletion of RhoGEF12 protects mice from overload-induced hypertrophy, fibrosis, and development of heart failure. Importantly, in mice with preexisting hypertrophy, induction of RhoGEF12 deficiency protects from cardiac decompensation, resulting in significantly increased long-term survival. Collectively, RhoGEF12 acts as an integrator of stretch-induced signaling cascades in cardiomyocytes and is an interesting new target for therapeutic intervention in patients with pressure overload–induced heart failure.
In response to increased pressure or volume load, for example in hypertension or valve disease, the heart undergoes hypertrophy, an initially compensatory response which, if the stimulus persists, may become maladaptive and result in chronic heart failure (Frey et al., 2004; Hill and Olson, 2008). Mechanical stress is regarded as the primary stimulus for cardiac remodeling, and several mechanosensitive structures have been suggested to translate changes in physical force into intracellular signals, for example ion channels, sarcomeric proteins, or integrins (Sadoshima and Izumo, 1997; Lammerding et al., 2004; Brancaccio et al., 2006). In addition to these direct sensors of stretch, various locally or systemically released humoral factors have been implicated in the hypertrophic response, for example growth factors or agonists at G protein–coupled receptors (GPCRs; Ito et al., 1993; Sadoshima et al., 1993; Sadoshima and Izumo, 1997; Rockman et al., 2002; Dorn and Hahn, 2004). Together, these signaling pathways converge on a limited number of intracellular signaling cascades including mitogen-activated protein kinases, the PI3K–Akt–GSK-3 pathway, calcium/calmodulin-dependent calcineurin phosphorylation, or small GTPases such as Ras, Rac, or RhoA (Clerk and Sugden, 2000; Frey and Olson, 2003; Heineke and Molkentin, 2006; Miyamoto et al., 2010). RhoA is a molecular switch that cycles between an inactive, GDP-bound state and an active, GTP-bound state; it controls various cellular functions related to the actin cytoskeleton, including cell shape, migration, adhesion, and transcriptional regulation (Hall, 1998; Clerk and Sugden, 2000; Olson and Nordheim, 2010). GTP binding to RhoA is stimulated by Rho guanine nucleotide exchange factors (RhoGEFs; Rossman et al., 2005), which in turn can be activated by integrins, receptor tyrosine kinases, and heterotrimeric G proteins of the families Gi, Gq/11, and G12/13 (Burridge and Wennerberg, 2004). By what mechanisms RhoA is activated in adult cardiomyocytes under conditions of pressure overload, which downstream effectors it controls, and whether these pathways are relevant for cardiac remodeling in vivo are currently unclear.
RESULTS AND DISCUSSION
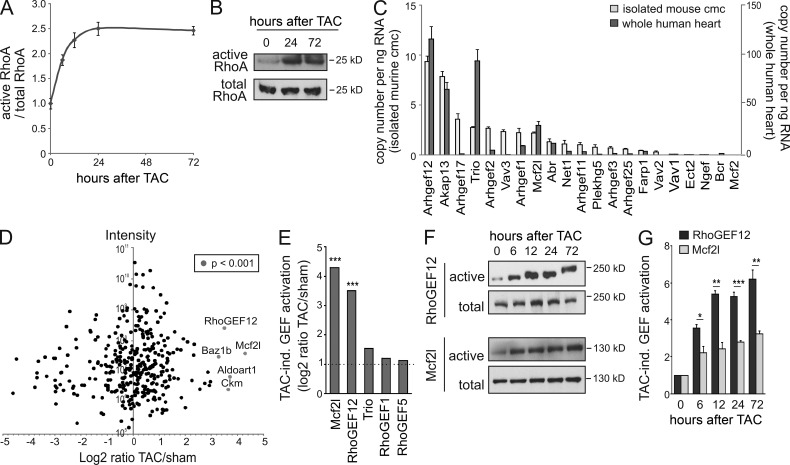
To impose pressure overload on left cardiac ventricles in vivo, we used transverse aortic constriction (TAC) in mice, which resulted in a rapid and sustained RhoA activation (Fig. 1, A and B). Quantitative RT-PCR (qRT-PCR) revealed that both adult murine cardiomyocytes and whole human hearts expressed various RhoGEFs, most abundant among them Arhgef12, the gene coding for RhoGEF12 (also known as leukemia-associated RhoGEF [LARG]; Fig. 1 C). To investigate which of these RhoGEFs were activated in the mouse heart by TAC, we performed affinity pull-down assays with a nucleotide-free RhoA mutant, a method designed to precipitate active pools of Rho-interacting proteins (Garcia-Mata et al., 2006), followed by mass spectrometry (MS). The two RhoGEFs that showed strongest activation in response to TAC were Mcf2l and RhoGEF12, whereas other RhoGEFs did not show significantly increased RhoA binding after TAC (Fig. 1, D and E). Protein immunoblotting of left ventricular lysates obtained at different time points after TAC confirmed the strong and sustained activation of RhoGEF12, whereas activation of Mcf2l was less prominent (Fig. 1, F and G).
Figure 1.
Pressure overload–induced RhoGEF activation in vivo. (A and B) RhoA activation in wild-type hearts at different time points after TAC was determined by ELISA (n = 6; A) or by pull-down assay and consecutive Western blotting (n = 2; B). (C) qRT-PCR analysis of RhoGEF expression in isolated adult murine cardiomyocytes (cmc; n = 3) and whole in human hearts (n = 2). (D and E) Activation of RhoGEF proteins 24 h after TAC was determined by mass spectrometric analysis of proteins coprecipitated with bead-coupled nucleotide-free RhoA. D shows the label-free ratio distribution against the sum of peak intensities, and E depicts the five RhoGEFs that showed at least twofold increase in RhoA binding after TAC (n = 1; data presented as log2 of the LFQ intensity ratio between TAC sample and sham sample, and significance indicates ratio outliers from the main distribution). (F) Activation of RhoGEF12 and Mcf2l was determined at different time points after TAC by precipitating RhoA-interacting proteins with bead-coupled nucleotide-free RhoA mutant as in D, followed by immunoblotting with antibodies directed against Mcf2l and RhoGEF12 (total cell lysate as loading control; n = 3). (G) Statistical evaluation of F (basal set to 1). Error bars indicate SEM. *, P <0.05; **, P < 0.01; ***, P < 0.001.
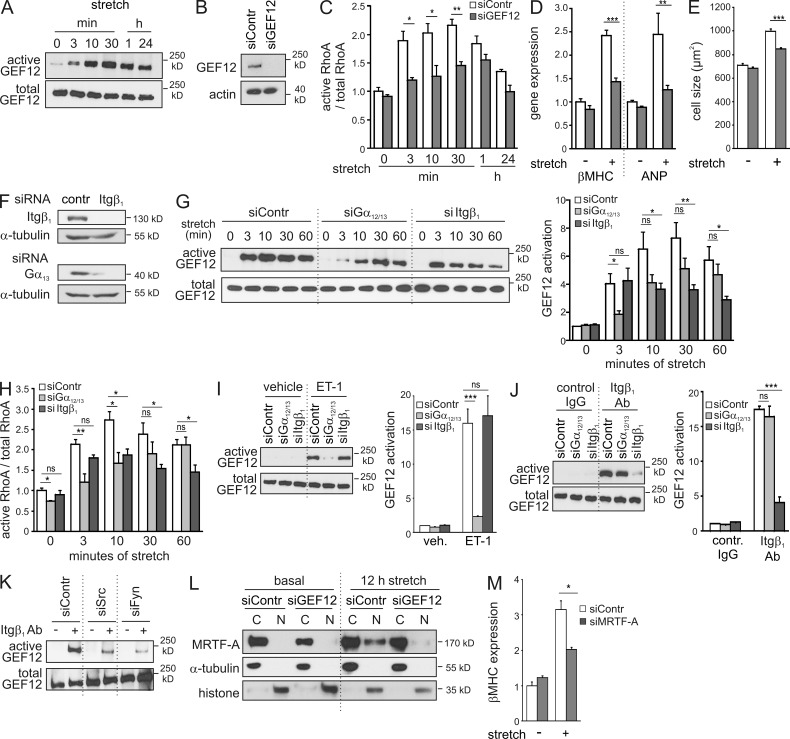
To investigate the role of RhoGEF12-dependent RhoA activation in cardiomyocyte hypertrophy, we studied stretch-induced effects in cultured neonatal rat ventricular myocytes (NRVMs) in vitro. Mechanical stress induced a fast and stable activation of RhoGEF12 (Fig. 2 A) and RhoA (Fig. 2 C) with a maximal response between 3 and 30 min. siRNA-mediated knockdown of RhoGEF12 (Fig. 2 B) strongly reduced stretch-induced RhoA activation (Fig. 2 C) as well as expression of hypertrophy-specific genes such as β-MHC or atrial natriuretic peptide (ANP; Fig. 2 D). Also, stretch-induced increases in cell size were significantly reduced after knockdown of RhoGEF12 (Fig. 2 E). Pretreatment of NRVMs with the RhoA inhibitor C3 exoenzyme or siRNA-mediated knockdown of RhoA fully mimicked the effect of RhoGEF12 knockdown (not depicted), indicating that RhoGEF12 controls hypertrophic gene expression through RhoA activation. We next studied the role of potential activators of RhoGEF12 such as integrin β1 (Itgβ1; Guilluy et al., 2011), G12/13 (Fukuhara et al., 2000), Gq/11 (Booden et al., 2002), or CD44 (Bourguignon et al., 2006). siRNA-mediated knockdown in NRVMs (Fig. 2 F) revealed that inactivation of Itgβ1 or the α-subunits of the G12/13 family (Gα12/13) time-dependently reduced stretch-induced RhoGEF12 activation (Fig. 2 G), and comparable effects were observed on the level of RhoA activation (Fig. 2 H). In contrast, knockdown of Gαq/11, CD44, or Itgβ3 was without effect (not depicted). We next investigated whether Itgβ1 and G proteins of the G12/13 family are independent activators of RhoGEF12 or whether they act in a sequential manner. We found that RhoGEF12 activation in response to endothelin-1 (ET-1), an agonist known to activate G12/13-coupled GPCRs but not integrins, depended on Gα12/13 but not on Itgβ1 (Fig. 2 I), whereas RhoGEF12 stimulation induced by an Itgβ1-activating antibody required Itgβ1 but not Gα12/13 (Fig. 2 J). We furthermore found that knockdown of protein kinases c-Src and Fyn strongly reduced RhoGEF12 activation elicited by direct Itgβ1 stimulation (Fig. 2 K), suggesting that these kinases mediate Itgβ1-dependent RhoGEF activation in cardiomyocytes. With respect to the mechanisms linking RhoA activation to transcriptional regulation, we studied the potential involvement of myocardin-related transcription factors (MRTFs), which have been shown to translocate upon RhoA-mediated actin polymerization to the nucleus where they act as coactivators of serum response factor (SRF)–dependent gene transcription (Olson and Nordheim, 2010). We found that stretch-induced translocation of MRTF-A from the cytoplasmic fraction to the nuclear fraction depended on RhoGEF12 (Fig. 2 L) and that stretch-induced expression of hypertrophy-specific genes such as β-MHC depended on MRTF-A (Fig. 2 M).
Figure 2.
Mechanism of stretch-induced RhoGEF12 activation in NRVMs. (A) Stretch-induced RhoGEF12 activation was determined by precipitating RhoA-interacting proteins with a bead-coupled nucleotide-free RhoA mutant, followed by immunoblotting with antibodies directed against RhoGEF12 (n = 3). (B) Efficiency of RhoGEF12 knockdown in NRVMs as shown by immunoblotting with anti-RhoGEF12 antibodies (actin as loading control; n = 2). (C) RhoA activation in siRNA-treated NRVMs at different time points after exposure to cyclic stretch (n = 3; basal set to 1). (D) Expression of hypertrophy-specific genes in siRNA-transfected NRVMs was determined by qRT-PCR after 24 h of cyclic stretching (n = 3; data normalized to GAPDH, control values set to 1). (E) Effect of RhoGEF12 (GEF12) knockdown on cell size in the basal state and after 24 h of cyclic stretching (n = 2). (F) Efficiency of siRNA-mediated knockdown of Itgβ1 or Gα13 in NRVMs (α-tubulin as loading control; n = 2). (G) Stretch-induced activation of RhoGEF12 after knockdown of Gα12/13 or Itgβ1 was determined by affinity pull-down assay with nucleotide-free RhoA and consecutive immunoblotting (n = 8; basal set to 1; data in B–G were generated using two independent sets of siRNAs). (H) Stretch-induced RhoA activation was determined in NRVMs after siRNA-mediated knockdown of Gα12/13 or Itgβ1 (n = 4). (I and J) RhoGEF12 activation in response to 1 µM ET-1 (5 min; I) or in response to 5 µg/ml of an activating Itgβ1 antibody (Ab; 5 min; J) in siRNA-treated NRVMs (left, exemplary immunoblot; right, statistical evaluation; n = 3; basal set to 1). (K) RhoGEF12 activation induced by an activating Itgβ1 antibody after siRNA-mediated knockdown of Src (siSrc) or Fyn (siFyn; n = 3). (L) Translocation of MRTF-A from the cytosolic fraction (C) to the nuclear fraction (N) in siRNA-transfected NRVMs after 12 h of stretch. Antibodies against tubulin and histone were used as markers for cytosolic and nuclear fractions, respectively (n = 2). (M) Effect of siRNA-mediated knockdown of MRTF-A on stretch-induced up-regulation of β-MHC expression in NRVMs (n = 3). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
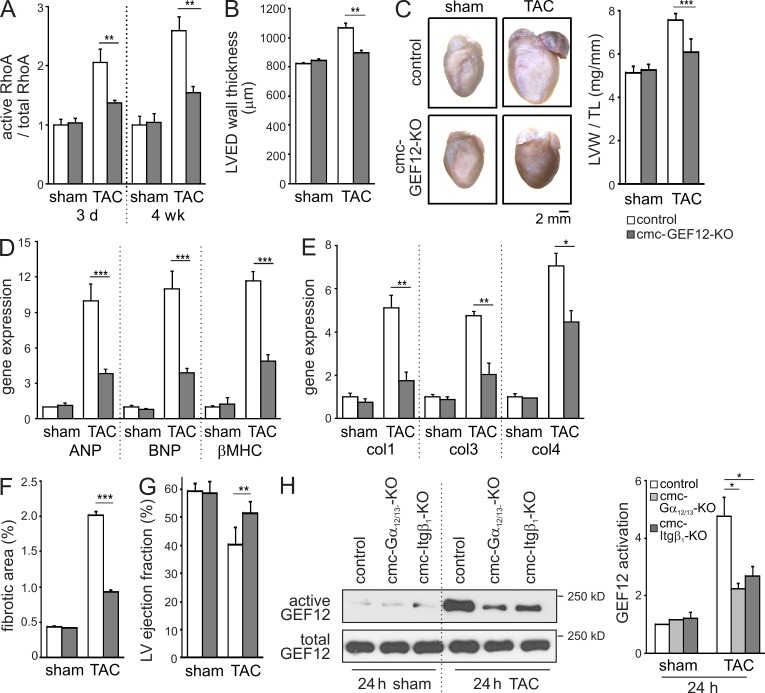
To study the role of RhoGEF12 in cardiomyocytes in vivo, we generated mice with tamoxifen-inducible, cardiomyocyte-specific RhoGEF12 deficiency (cmc-GEF2-KO). Magnetic resonance imaging (MRI) and histological analyses did not reveal basal differences between the genotypes (not depicted). When pressure overload was induced by TAC, cmc-GEF12-KOs showed reduced RhoA activation compared with control mice (Fig. 3 A). 4 wk after TAC, the left ventricular wall thickness (Fig. 3 B), left ventricular weight/tibia length ratio (Fig. 3 C), and expression of hypertrophy-specific genes (Fig. 3 D) were significantly reduced in mutant mice. What is more, cmc-GEF12-KOs showed lower expression of collagen isoforms (Fig. 3 E) and reduced fibrosis (Fig. 3 F), and the left ventricular ejection fraction was preserved compared with control mice (Fig. 3 G). To investigate whether Gα12/13 and Itgβ1 are also under in vivo conditions relevant for RhoGEF12 activation, we studied TAC-induced RhoGEF12 activation in mice with tamoxifen-inducible, cardiomyocyte-specific Gα12/13 deficiency (cmc-Gα12/13-KO; Takefuji et al., 2012) and tamoxifen-inducible, cardiomyocyte-specific Itgβ1 deficiency (cmc-Itgβ1-KO). We found that in both mutant mouse lines, activation of RhoGEF12 was significantly reduced 24 h after TAC, indicating that also under conditions of pressure overload Itgβ1 and Gα12/13 are crucial for RhoGEF12 activation (Fig. 3 H).
Figure 3.
RhoGEF12 mediates pressure overload–induced hypertrophy in vivo. (A) RhoA activation in adult hearts from control mice and cmc-GEF12-KOs 3 d and 4 wk after TAC or sham surgery (n = 3). (B) Left ventricular end-diastolic (LVED) wall thickness (as determined by MRI) 4 wk after sham operation or TAC (n = 6–7). (C) Cardiac morphology and left ventricular weight/tibia length (LVW/TL) ratio 4 wk after TAC or sham surgery (n = 8–12). (D and E) Expression of hypertrophy-specific genes (D) and profibrotic genes (E) was determined by qRT-PCR in hearts of control mice and cmc-GEF12-KOs 4 wk after sham or TAC (n = 4–6). Col1, 3, and 4: collagen isoforms 1, 3, and 4. (F) TAC-induced fibrosis determined by Picrosirius red staining 4 wk after TAC (n = 4–6). (G) Left ventricular (LV) ejection fraction determined by MRI 4 wk after sham surgery or TAC (n = 6–7). (H) TAC-induced RhoGEF12 activation in hearts of control mice as well as in hearts of tamoxifen-inducible, cardiomyocyte-specific Gα12/13-deficient mice (cmc-Gα12/13-KO) and tamoxifen-inducible, cardiomyocyte-specific Itgβ1-deficient mice (cmc-Itgβ1-KO) was determined by precipitating RhoA-interacting proteins with a bead-coupled nucleotide-free RhoA mutant, followed by immunoblotting with antibodies directed against RhoGEF12 (n = 2). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
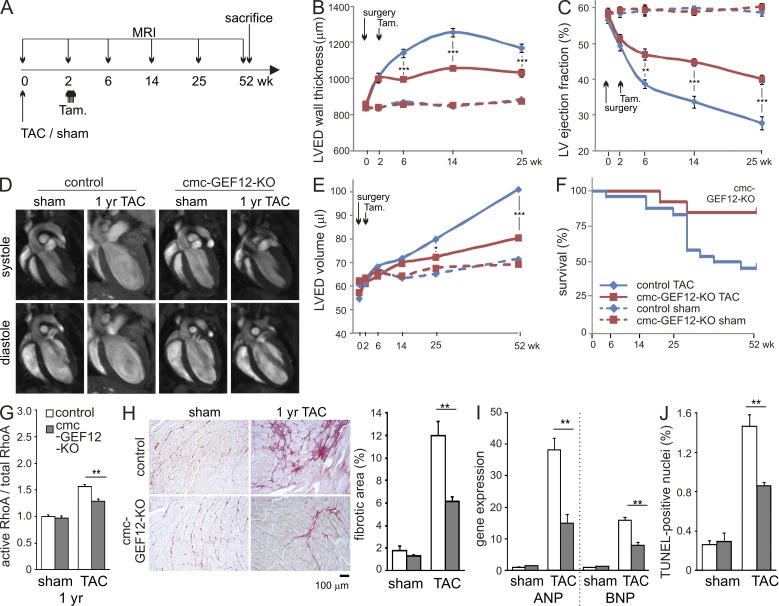
Based on the finding that inactivation of RhoGEF12 in cardiomyocytes prevents not only the development of hypertrophy but also deterioration of cardiac ejection fraction, we next investigated whether inhibition of RhoGEF12 would also prove beneficial if applied in already established hypertrophy. To study this, TAC was performed 2 wk before tamoxifen-mediated induction of RhoGEF12 deficiency, and cardiac performance was followed by MRI for up to 1 yr (Fig. 4 A). Both genotypes behaved similarly during the first 2 wk after surgery, but upon tamoxifen treatment, RhoGEF12-deficient mice were protected from further increases in wall thickness and deterioration of ejection fraction (Fig. 4, B and C). Interestingly, at ∼6 mo after TAC, wild-type mice but not cmc-GEF12-KOs started to show progressive ventricular dilation (Fig. 4, D and E), resulting in significantly increased mortality (Fig. 4 F). Hearts of surviving control mice 1 yr after TAC compared with mutants showed significantly higher RhoA activation, stronger fibrosis, elevated expression of heart failure markers ANP and brain natriuretic peptide (BNP), and increased apoptosis (Fig. 4, G–J).
Figure 4.
Cardiomyocyte-specific inactivation of RhoGEF12 improves cardiac function and survival in preexisting hypertrophy. (A) Experimental design. (B–E) Control mice (blue lines, diamonds) and not yet induced cmc-GEF12-KOs (red lines, squares) were subjected to sham surgery (dashed lines) or TAC (solid lines) at day 0, followed by tamoxifen (Tam) induction of recombination on days 14–18. MRI analysis of left ventricular end-diastolic (LVED) wall thickness (B), left ventricular (LV) ejection fraction (C), and LVED volume (D, exemplary MRI images; E, statistical evaluation) was performed before and 2, 6, 14, 25, and 52 wk after surgery (n = 15). (F) Survival plot for control and cmc-GEF12-KOs up to 1 yr after TAC, followed by tamoxifen injection (n = 18–24). (G) RhoA activation in adult hearts from control mice and cmc-GEF12-KOs 1 yr after TAC (n = 3). (H) TAC-induced fibrosis 1 yr after TAC as determined by Picrosirius red staining (n = 5). (I) Expression of ANP and BNP 1 yr after sham surgery or TAC as determined by qRT-PCR in whole hearts (n = 3–5; data normalized to GAPDH, control values set to 1). (J) Apoptosis was determined by TUNEL staining in left ventricles 1 yr after TAC (n = 3). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Collectively, we show in this study that RhoGEF12 integrates signals from integrins and GPCRs to control hypertrophic gene expression and the transition to heart failure. Because of their strategic position at the interface between extracellular matrix and intracellular cytoskeleton, integrins have repeatedly been suggested as sensors of mechanical stress (Wang et al., 2009). Evidence for a role of integrins in cardiac hypertrophy mainly stems from studies in NRVMs showing that cell growth and ANP production are reduced after blockade of β integrins (Yutao et al., 2006) and that stretch activates integrin-dependent signaling intermediates such as focal adhesion kinase, integrin-linked kinase, or Src (Laser et al., 2000; Torsoni et al., 2003; Lammerding et al., 2004; Brancaccio et al., 2006). Integrins are also well known for their ability to modulate RhoA activity (Burridge and Wennerberg, 2004), but the role of integrin-dependent RhoA activation in the heart is not understood. Our data show that mechanical stress induces Itgβ1-dependent RhoA activation in neonatal cardiomyocytes and that RhoGEF12 is the major RhoGEF contributing to this effect. In line with this, it was recently shown that application of force to β integrins induces in fibroblasts recruitment of RhoGEF12 to focal adhesions, resulting in RhoA activation (Guilluy et al., 2011). The same study identified Arhgef2/GEF-H1 as a second GEF involved in force-induced RhoA activation, but our mass spectrometric analysis suggested that TAC-induced Arhgef2/GEF-H1 activation in adult cardiomyocytes is mild compared with RhoGEF12 activation.
Our data furthermore show that stretch-induced RhoGEF12 activation is not only mediated by Itgβ1, but also requires heterotrimeric G proteins of the G12/13 family, whereas the Gq/11 family seems dispensable. The stretched myocardium is known to release several humoral factors that enhance the hypertrophic response through stimulation of GPCRs, for example ET-1 or angiotensin II (Ito et al., 1993; Sadoshima et al., 1993), and so far these effects were generally believed to be mediated by the Gq/11 family (Frey and Olson, 2003; Dorn and Hahn, 2004). However, the receptors in question can also signal through the G12/13 family (Riobo and Manning, 2005), and our findings clearly suggest a role of G12/13 in stretch-induced hypertrophy in vitro. This notion is in line with several in vitro studies in NRVMs (Miyamoto et al., 2010) and is furthermore supported by the fact that TAC-induced hypertrophy is strongly reduced in cardiomyocyte-specific G12/13-deficient mice (Takefuji et al., 2012). Interestingly, G12/13 seemed mainly responsible for early stretch-induced RhoGEF12 activation, whereas Itgβ1 was required for later effects, together creating a unique temporal pattern of RhoA activation in response to stretch.
The mechanisms underlying the transition from hypertrophy to heart failure are not well defined, but cardiomyocyte apoptosis and fibrotic replacement are most likely contributing factors (Dorn and Hahn, 2004; Hill and Olson, 2008; Miyamoto et al., 2010). Our study reveals that inactivation of one particular path of RhoA activation, namely RhoGEF12-dependent RhoA activation, protects the heart not only from hypertrophy development, but also from cardiomyocyte apoptosis, fibrosis, and the development of chronic heart failure. RhoA itself is at the center of several signaling pathways (Burridge and Wennerberg, 2004), and its role in cardiac physiology and pathophysiology is correspondingly complex: although strong RhoA activation in cardiomyocytes has been shown to induce apoptosis and cardiomyopathy (Miyamoto et al., 2010), moderate RhoA activation has cardioprotective effects under conditions of ischemia/reperfusion (Xiang et al., 2011). Whether a general inhibition of RhoA activation would have the same beneficial effects during pressure overload as prevention of RhoGEF12-dependent RhoA activation is currently unclear.
In conclusion, we found that RhoGEF12 plays a unique role in mediating stretch-induced cellular responses by integrating signals from activated GPCRs and integrins. The fact that genetic ablation of RhoGEF12 does not affect basal heart function but efficiently prevents cardiac decompensation even in mice with established hypertrophy makes this signaling pathway a promising target for therapeutic intervention in patients with pressure overload–induced heart failure.
MATERIALS AND METHODS
Materials and chemicals.
ET-1 and tamoxifen were purchased from Sigma-Aldrich. C3-exoenzyme (cell-permeable Rho inhibitor [CT03]) was obtained from Cytoskeleton. Antibodies to Gα13 (A-20), RhoGEF12 (LARG; N14), Mcf2l (Dbs; L-20), Histone (N-16), MRTF-A (C-19), and β-actin (C-4) were purchased from Santa Cruz Biotechnology, Inc. Activating Itgβ1 antibodies (clone 18/CD29) were obtained from BD, and α-tubulin antibodies were obtained from Sigma-Aldrich.
Experimental animals.
The generation of Arhgef12fl/fl mice and αMHC-CreERT2+/− mice has been reported previously (Herroeder et al., 2009; Takefuji et al., 2012). Cre-mediated recombination of floxed alleles was induced by intraperitoneal injection of 1 mg tamoxifen dissolved in 50 µl Miglyol on five consecutive days. Vehicle-treated mice received Miglyol only. For in vivo experiments, tamoxifen-treated α-MHC–CreERT2+/−; Arhgef12wt/wt mice were used as controls. Experiments were performed 2 wk after the end of induction and were approved by local authorities (Regierungspräsidium Darmstadt, Hessen).
TAC.
For TAC, male mice aged 8–10 wk were anesthetized with 50 mg/kg pentobarbital sodium. After intubation, the chest was opened and the aortic arch was identified. TAC was created by ligating the transverse aorta between the right innominate and the left common carotid artery against a blunted 24-gauge needle using a 7-0 suture. The needle was then gently retracted. The sham procedure was identical except that the aorta was not ligated.
Determination of activated RhoGEF12.
Determination of activated RhoGEF12 was performed as described previously (García-Mata et al., 2006) with the following modifications: Hearts were extirpated and frozen in liquid nitrogen and then disrupted by a homogenizer in lysis buffer (0.2% Triton X-100, 20 mM Hepes, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and protease inhibitors), and the protein concentration of the supernatants was determined after centrifugation by Precision Red Advanced Protein Assay Reagent (Cytoskeleton). Samples containing 1.5 mg of total protein were incubated at 4°C for 1 h with 20 µg GST-RhoAG17A bound to Glutathione–Sepharose 4B beads (GE Healthcare). The beads were washed four times with lysis buffer and then eluted with SDS sample buffer. The eluates were subjected to immunoblot analysis with an anti-RhoGEF12 antibody.
Determination of active and total RhoA.
Activated RhoA and total RhoA were measured by G-LISA RhoA Activation Assay kit and Total RhoA ELISA kit (both from Cytoskeleton) using 0.6 mg/ml protein per sample. For the analysis of active and total RhoA in left ventricles after TAC, hearts were extirpated and snap-frozen in liquid nitrogen. 50 mg of left ventricular tissue was disrupted using a homogenizer in the lysis buffer provided with the G-LISA RhoA Activation Assay kit. The lysates were centrifuged, and the supernatants were snap-frozen in liquid nitrogen. After examining the total protein concentration in supernatants by Precision Advanced Protein Assay Reagent (Cytoskeleton), supernatants were mixed with ice-cold binding buffer for RhoA activation assay and with sample dilution buffer for total RhoA assay. Samples were then pipetted into RhoA activation assay plates and total RhoA assay plates, respectively. After incubation, the plates were washed with washing buffer and incubated with anti-RhoA primary antibody. After washing, the plates were incubated with secondary HRP-labeled antibody. After washing five times, the plates were incubated with color development reagent and read by a microplate spectrophotometer (OD 490 nm; Multiskan Spectrum; Thermo Fisher Scientific). Data were expressed as a ratio of active RhoA/total RhoA; the basal values of the control group were set to 1.
RhoA pull-down assay.
Hearts were extirpated and frozen in liquid nitrogen. 50 mg of left ventricular tissue was disrupted using a homogenizer in 500 µl of lysis buffer (0.2% Triton X-100, 20 mM Hepes, pH 7.5, 0.1 mM EGTA, 10 mM MgCl2, 500 mM NaCl, and protease inhibitors) containing 30 µg GST-Rho–binding domain of Rhotekin (GST-RBD). The lysates were centrifuged and the supernatants were incubated with Glutathione–Sepharose 4B beads for 60 min at 4°C. The beads were washed four times with lysis buffer and then eluted with SDS sample buffer. The eluates were subjected to SDS-PAGE, followed by immunoblotting with an anti-RhoA antibody (Santa Cruz Biotechnology, Inc.).
Isolation of neonatal rat ventricular cardiomyocytes.
NRVMs were isolated from 1–2-d-old rat neonates using a kit from Worthington Biochemical Corporation. After digestion, cells were preplated for 1 h to remove nonmyocytes, plated on cell culture dishes, and then cultured in DMEM with 10% fetal bovine serum. The next day, cells were cultured in serum-free DMEM containing 100 µM BrdU (Sigma-Aldrich). NRVMs were transfected with siRNAs (QIAGEN or Sigma-Aldrich) using Lipofectamine RNAiMAX (Invitrogen) 3 and 20 h after plating according to the manufacturer’s instructions. The following siRNA target sequences were used: RhoGEF12-I, 5′-GTCTCAAGTTGTCTGAGTA-3′; RhoGEF12-II, 5′-CCAAGTATTCTATCAGCGA-3′; ITGB-1-I, 5′-CATTGGAGATGAGGTTCAA-3′; ITGB-1-II, 5′-GTGATAACTTCAACTGCGA-3′; Gα13-I, 5′-CAGCAACGTGATCAAAGGTAT-3′; Gα13-II, 5′-CAGTATCTTCCTGCTATAAGA-3′; Gα12-I, 5′-CCGCGACACCATCTTCGACAA-3′; Gα12-II, 5′-GTGAGTCAGTGAAGTACTT-3′; ITGB-3, 5′-GCTTTGACGCCATCATGCA-3′; Gαq, 5′-AAGCACTCTTTAGAACCATTA-3′; Gα11, 5′-CACAACTGGCATCATCGAGTA-3′; CD44, 5′-CTACTTCACTGGAAGGCTA-3′; FYN, 5′-GAGAATCCCTGCAGTTGAT-3′; SRC, 5′-CAGCTTGTGGCTTACTACT-3′; RhoA, 5′-CAGACACTGATGTTATACT-3′; and Mkl1 (MRTF-A), 5′-CAATTTGCCTCCACTTAGT-3′.
Isolation of adult mouse left ventricular cardiomyocyte.
Adult mouse cardiomyocytes were isolated as previously described (Takefuji et al., 2012). In brief, the heart was removed quickly and cannulated from the aorta with a blunted 27-gauge needle to allow retrograde perfusion of the coronary arteries. The heart was first washed with 50 ml of perfusion buffer (113 mM NaCl, 4.7 mM KCl, 0.6 mM KH2PO4, 1.2 mM MgSO4, 12 mM NaHCO3, 10 mM KHCO3, 10 mM Hepes, 30 mM taurine, 10 mM 2,3-Butanedione monoxime, and 5.5 mM glucose, pH 7.46) for 10 min and then digested with 75 ml of digesting buffer (perfusion buffer with 0.05 mg/ml Liberase DH [Roche] and 12.5 µM CaCl2) for 20 min. The heart was removed from the perfusion apparatus, and the left ventricle was minced with a forceps in digesting buffer. Centrifugation (50 g, 1 min) was performed three times to enrich cardiomyocytes.
Cell culture.
To study the cellular effects of stretch, NRVMs were incubated in serum-free medium for 18 h and then stretched by 10% with a frequency of 1 Hz using a Flexcell system. Stretch-induced changes in cardiomyocyte size were quantified using National Institutes of Health ImageJ software; 75 cells were evaluated per experimental condition. To study the cellular effects of direct stimulation of G12/13 signaling or Itgβ1 signaling, NRVMs were seeded on 6-well tissue culture plates without coating (Greiner Bio-One). NRVMs were incubated in serum-free medium for 18 h and then incubated in 500 µl of serum-free medium containing IgG, 5 µg/ml of a stimulating anti-Itgβ1 antibody (clone 18/CD29; BD), or 1 µM ET-1 for 5 min. After 5-min incubation, RhoGEF pull-down assay was performed. To study stretch-induced MRTF-A translocation, isolation of nuclear protein and cytoplasmic protein extraction was performed with DUALXtract (Dualsystems Biotech).
Western blotting.
Samples were subjected to SDS-PAGE, transferred to nitrocellulose transfer membranes (Whatman), and then incubated with primary antibodies as indicated. Equal loading was checked using antibodies to α-tubulin or actin.
Histological analyses.
Freshly dissected heart tissue was fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Embedded hearts were stained with Picrosirius red according to standard protocols. 20 randomly chosen frames from the sections were quantified to assess the degree of heart fibrosis using ImageJ software. TUNEL staining of nuclei positive for DNA strand breaks was performed using the Cell Death Detection kit (Roche) according to the manufacturer’s instructions. In brief, paraffin-embedded sections were deparaffinized and rehydrated. The sections were then incubated with Proteinase K at room temperature for 30 min. The sections were incubated with TUNEL reaction mixture at 37°C for 1 h. Nuclear density was determined by manual counting of DAPI-stained nuclei. At least 10,000 DAPI-positive cells were counted in each animal.
Mass spectrometric analysis.
After immunoprecipitation using bead-coupled GST-RhoAG17A (see section “Determination of activated RhoGEF12”) from sham- and TAC-operated hearts, proteins were separated by one-dimensional SDS-PAGE (4–12% Novex gels; Invitrogen) and stained with colloidal Coomassie. Gel bands were excised and subjected to in gel digest with trypsin. The resulting tryptic peptides were extracted with acetonitrile and desalted with reversed phase C18 STAGE tips (Luber et al., 2010). Mass spectrometric experiments were performed on a nanoflow HPLC system (Agilent Technologies) connected to an LTQ-Orbitrap instrument (Thermo Fisher Scientific) equipped with a nanoelectrospray source (Proxeon). The mass spectrometer was operated in the data-dependent mode to monitor MS and MS/MS spectra. Survey full-scan MS spectra (from m/z 300–2,000) were acquired in the Orbitrap with a resolution of R = 60,000 at m/z 400 after accumulation of 1,000,000 ions. The five most intense ions from the preview survey scan delivered by the Orbitrap were sequenced by collision-induced dissociation in the LTQ. Mass spectra were analyzed using MaxQuant software (version 1.2.2.9; Luber et al., 2010), and all tandem mass spectra were searched against the mouse and rat International Protein Index protein sequence database (mouse IPI version 3.68) and concatenated with reversed copies of all sequences. The required false positive rate was set to 1% at the protein and peptide level. Maximum allowed mass deviation was set to 20 ppm in MS mode and 0.5 D for MS/MS peaks. Cysteine carbamidomethylation was searched as a fixed modification, and N-acetyl protein and oxidized methionine were searched as variable modifications. A maximum of three missed cleavages were allowed. Protein quantitation was performed with the MaxQuant label-free option (Luber et al., 2010). Data are presented as the log2 of the LFQ intensity ratio between TAC-operated sample and sham-operated sample. Significance was calculated with the MaxQuant software tool and indicates ratio outliers from the main distribution.
mRNA expression analysis.
RNA was extracted from left ventricles with the RNA fibrosis tissue kit (QIAGEN) and from NRVMs or adult mouse cardiomyocytes with the RNeasy Mini kit (QIAGEN) according to the manufacturer’s protocol. RT reaction was performed using the QuantiTect Reverse Transcription kit (QIAGEN). Quantification of human and mouse genes was performed using the LightCycler 480 Probe Master System (Roche; for primers and probes, see Tables S1 and S2). Pooled human cardiac RNAs were obtained from Takara Bio Inc. (Caucasian males, 30–39 yr of age, cause of death: trauma). Genomic DNA from mouse tails or HUVECs was used as a universal standard to calculate gene copy number per nanogram of RNA. For hypertrophy-specific genes and fibrosis genes, data are presented after normalization to GAPDH, and basal values were set to 1. qRT-PCR for rat genes was performed using the LightCycler 480 SYBR Green Master (Roche; for primer sequences, see Table S3).
MRI.
Cardiac MRI measurements were performed on a 7.0 T Bruker Pharmascan, equipped with a 300 mT/m gradient system, using a custom-built circularly polarized birdcage resonator and the Early Access Package for self-gated cardiac Imaging (Bruker; Takefuji et al., 2012). The mice were measured under volatile isoflurane (2.0%) anesthesia. The measurement is based on the gradient echo method (repetition time = 6.2 ms; echo time = 1.6 ms; field of view = 2.20 × 2.20 cm; slice thickness = 1.0 mm; matrix = 128 × 128; repetitions = 100). The imaging plane was localized using scout images showing the two- and four-chamber view of the heart, followed by acquisition in short axis view, orthogonal on the septum in both scouts. Multiple contiguous short-axis slices consisting of 9 or 10 slices were acquired for complete coverage of the left ventricle. MRI data were analyzed using Qmass digital imaging software (Medis).
Generation of tamoxifen-inducible, cardiomyocyte-specific KOs for Gα12/13 (cmc-Gα12/13-KO) and Itgβ1 (cmc-Itgβ1-KO).
The generation and characterization of cmc-Gα12/13-KOs has been described previously (Takefuji et al., 2012). cmc-Itgβ1-KOs were generated by intercrossing the tamoxifen-inducible, cardiomyocyte-specific α-MHC–CreERT2 line to mice carrying a floxed Itgβ1 allele (B6;129-Itgb1tm1Efu/J; The Jackson Laboratory). In both mouse lines, Cre-mediated recombination of floxed alleles was induced by intraperitoneal injection of 1 mg tamoxifen dissolved in 50 µl Miglyol on five consecutive days. Vehicle-treated mice received Miglyol only; α-MHC–CreERT2+/− mice not carrying floxed alleles were used as controls. Experiments were performed 2 wk after the end of induction.
Statistical analyses.
Data are presented as means ± SEM. Comparisons between two groups were performed with unpaired Student’s t test, and comparisons between more than two groups were performed by ANOVA followed by Bonferroni post hoc test. Survival curves were analyzed using Kaplan Meyer estimators and Log-rank (Mantel-Cox) test. Comparisons between more than two groups at different time points were performed by repeated measures ANOVA followed by Bonferroni post hoc test. “n” refers to the number of independent experiments or mice per group. P-values are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material.
Tables S1–S3 show primer sequences and probes used for qRT-PCR analyses. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122126/DC1.
Supplementary Material
Acknowledgments
We thank A. Wietelmann and U. Hofmann for introducing us to MRI imaging.
M. Takefuji was supported by a fellowship program of the Japan Research Foundation for Clinical Pharmacology.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANP
- atrial natriuretic peptide
- GPCR
- G protein–coupled receptor
- MRI
- magnetic resonance imaging
- MRTF
- myocardin-related transcription factor
- MS
- mass spectrometry
- NRVM
- neonatal rat ventricular myocyte
- qRT-PCR
- quantitative RT-PCR
- RhoGEF
- Rho guanine nucleotide exchange factor
- TAC
- transverse aortic constriction
References
- Booden M.A., Siderovski D.P., Der C.J. 2002. Leukemia-associated Rho guanine nucleotide exchange factor promotes G alpha q-coupled activation of RhoA. Mol. Cell. Biol. 22:4053–4061 10.1128/MCB.22.12.4053-4061.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Gilad E., Brightman A., Diedrich F., Singleton P. 2006. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J. Biol. Chem. 281:14026–14040 10.1074/jbc.M507734200 [DOI] [PubMed] [Google Scholar]
- Brancaccio M., Hirsch E., Notte A., Selvetella G., Lembo G., Tarone G. 2006. Integrin signalling: the tug-of-war in heart hypertrophy. Cardiovasc. Res. 70:422–433 10.1016/j.cardiores.2005.12.015 [DOI] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. 2004. Rho and Rac take center stage. Cell. 116:167–179 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- Clerk A., Sugden P.H. 2000. Small guanine nucleotide-binding proteins and myocardial hypertrophy. Circ. Res. 86:1019–1023 10.1161/01.RES.86.10.1019 [DOI] [PubMed] [Google Scholar]
- Dorn G.W., II, Hahn H.S. 2004. Genetic factors in cardiac hypertrophy. Ann. N. Y. Acad. Sci. 1015:225–237 10.1196/annals.1302.019 [DOI] [PubMed] [Google Scholar]
- Frey N., Olson E.N. 2003. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 65:45–79 10.1146/annurev.physiol.65.092101.142243 [DOI] [PubMed] [Google Scholar]
- Frey N., Katus H.A., Olson E.N., Hill J.A. 2004. Hypertrophy of the heart: a new therapeutic target? Circulation. 109:1580–1589 10.1161/01.CIR.0000120390.68287.BB [DOI] [PubMed] [Google Scholar]
- Fukuhara S., Chikumi H., Gutkind J.S. 2000. Leukemia-associated Rho guanine nucleotide exchange factor (LARG) links heterotrimeric G proteins of the G(12) family to Rho. FEBS Lett. 485:183–188 10.1016/S0014-5793(00)02224-9 [DOI] [PubMed] [Google Scholar]
- García-Mata R., Wennerberg K., Arthur W.T., Noren N.K., Ellerbroek S.M., Burridge K. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406:425–437 10.1016/S0076-6879(06)06031-9 [DOI] [PubMed] [Google Scholar]
- Guilluy C., Swaminathan V., Garcia-Mata R., O’Brien E.T., Superfine R., Burridge K. 2011. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 13:722–727 10.1038/ncb2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- Heineke J., Molkentin J.D. 2006. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7:589–600 10.1038/nrm1983 [DOI] [PubMed] [Google Scholar]
- Herroeder S., Reichardt P., Sassmann A., Zimmermann B., Jaeneke D., Hoeckner J., Hollmann M.W., Fischer K.D., Vogt S., Grosse R., et al. 2009. Guanine nucleotide-binding proteins of the G12 family shape immune functions by controlling CD4+ T cell adhesiveness and motility. Immunity. 30:708–720 10.1016/j.immuni.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Hill J.A., Olson E.N. 2008. Cardiac plasticity. N. Engl. J. Med. 358:1370–1380 10.1056/NEJMra072139 [DOI] [PubMed] [Google Scholar]
- Ito H., Hirata Y., Adachi S., Tanaka M., Tsujino M., Koike A., Nogami A., Murumo F., Hiroe M. 1993. Endothelin-1 is an autocrine/paracrine factor in the mechanism of angiotensin II-induced hypertrophy in cultured rat cardiomyocytes. J. Clin. Invest. 92:398–403 10.1172/JCI116579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Kamm R.D., Lee R.T. 2004. Mechanotransduction in cardiac myocytes. Ann. N. Y. Acad. Sci. 1015:53–70 10.1196/annals.1302.005 [DOI] [PubMed] [Google Scholar]
- Laser M., Willey C.D., Jiang W., Cooper G., IV, Menick D.R., Zile M.R., Kuppuswamy D. 2000. Integrin activation and focal complex formation in cardiac hypertrophy. J. Biol. Chem. 275:35624–35630 10.1074/jbc.M006124200 [DOI] [PubMed] [Google Scholar]
- Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O’Keeffe M., Mann M. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 32:279–289 10.1016/j.immuni.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Del Re D.P., Xiang S.Y., Zhao X., Florholmen G., Brown J.H. 2010. Revisited and revised: is RhoA always a villain in cardiac pathophysiology? J. Cardiovasc. Transl. Res. 3:330–343 10.1007/s12265-010-9192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Nordheim A. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo N.A., Manning D.R. 2005. Receptors coupled to heterotrimeric G proteins of the G12 family. Trends Pharmacol. Sci. 26:146–154 10.1016/j.tips.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Rockman H.A., Koch W.J., Lefkowitz R.J. 2002. Seven-transmembrane-spanning receptors and heart function. Nature. 415:206–212 10.1038/415206a [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Sadoshima J., Izumo S. 1997. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu. Rev. Physiol. 59:551–571 10.1146/annurev.physiol.59.1.551 [DOI] [PubMed] [Google Scholar]
- Sadoshima J., Xu Y., Slayter H.S., Izumo S. 1993. Autocrine release of angiotensin II mediates stretch-induced hypertrophy of cardiac myocytes in vitro. Cell. 75:977–984 10.1016/0092-8674(93)90541-W [DOI] [PubMed] [Google Scholar]
- Takefuji M., Wirth A., Lukasova M., Takefuji S., Boettger T., Braun T., Althoff T., Offermanns S., Wettschureck N. 2012. G(13)-mediated signaling pathway is required for pressure overload-induced cardiac remodeling and heart failure. Circulation. 126:1972–1982 10.1161/CIRCULATIONAHA.112.109256 [DOI] [PubMed] [Google Scholar]
- Torsoni A.S., Constancio S.S., Nadruz W., Jr, Hanks S.K., Franchini K.G. 2003. Focal adhesion kinase is activated and mediates the early hypertrophic response to stretch in cardiac myocytes. Circ. Res. 93:140–147 10.1161/01.RES.0000081595.25297.1B [DOI] [PubMed] [Google Scholar]
- Wang N., Tytell J.D., Ingber D.E. 2009. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10:75–82 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- Xiang S.Y., Vanhoutte D., Del Re D.P., Purcell N.H., Ling H., Banerjee I., Bossuyt J., Lang R.A., Zheng Y., Matkovich S.J., et al. 2011. RhoA protects the mouse heart against ischemia/reperfusion injury. J. Clin. Invest. 121:3269–3276 10.1172/JCI44371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutao X., Geru W., Xiaojun B., Tao G., Aiqun M. 2006. Mechanical stretch-induced hypertrophy of neonatal rat ventricular myocytes is mediated by beta(1)-integrin-microtubule signaling pathways. Eur. J. Heart Fail. 8:16–22 10.1016/j.ejheart.2005.05.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.