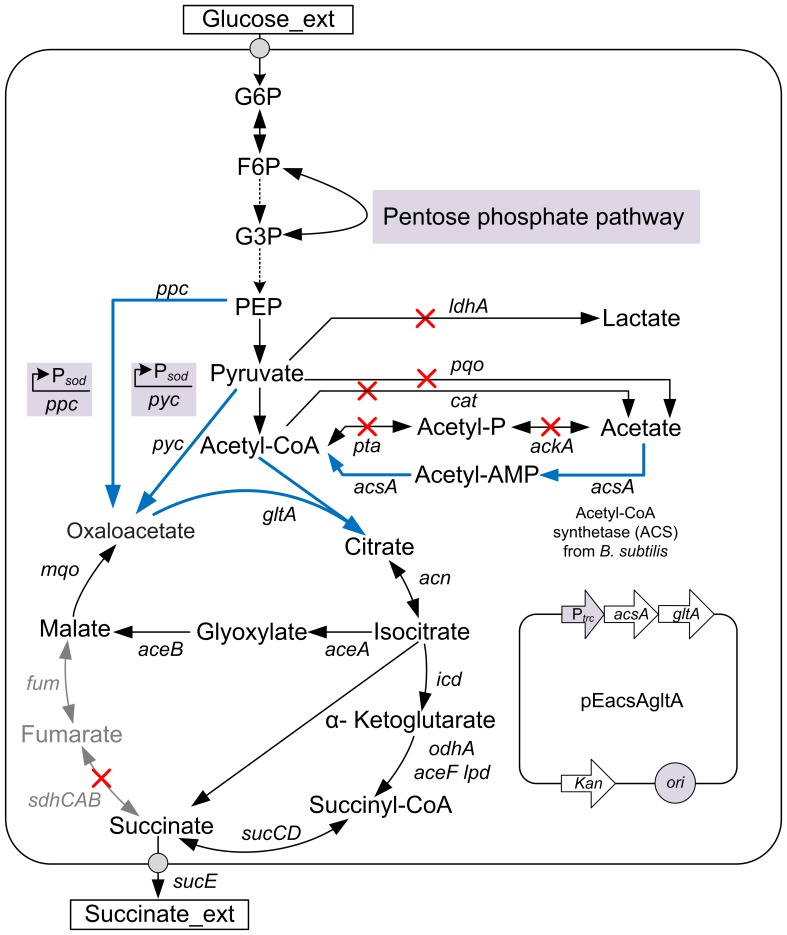
Figure 1. The central metabolic pathways of C. glutamicum ATCC 13032 and the engineering strategies for aerobic succinate overproduction.
Intermediate pathways omitted in glycolysis were illustrated by broken line. The red X indicated that the pathways were disrupted. The blue arrows indicated that pathways were overexpressed. Relevant reactions were represented by the genes encoding for corresponding enzymes: ppc, phosphoenolpyruvate carboxylase; pyc, pyruvate carboxylase; mqo, malate:quinone oxidoreductase; fum, fumarate hydratase; sdhCAB, succinate dehydrogenase complex; aceF lpd odhA, 2-oxoglutarate dehydrogenase complex; sucCD, succinyl-CoA synthetase, icd, isocitrate dehydrogenase; acn, aconitate hydratase; gltA, citrate synthase; aceA, isocitrate lyase; aceB, malate synthase; ldhA, L-lactate dehydrogenase; pqo, pyruvate : quinone oxidoreductase; pta, phosphotransacetylase; ackA, acetate kinase; cat, acetyl-CoA:CoA transferase; acsA, acetyl-CoA synthase; sucE, succinate exporter. Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; Acetyl-P, acetylphosphate; Acetyl-AMP, acetyladenylate; PEP, phosphoenolpyruvate.