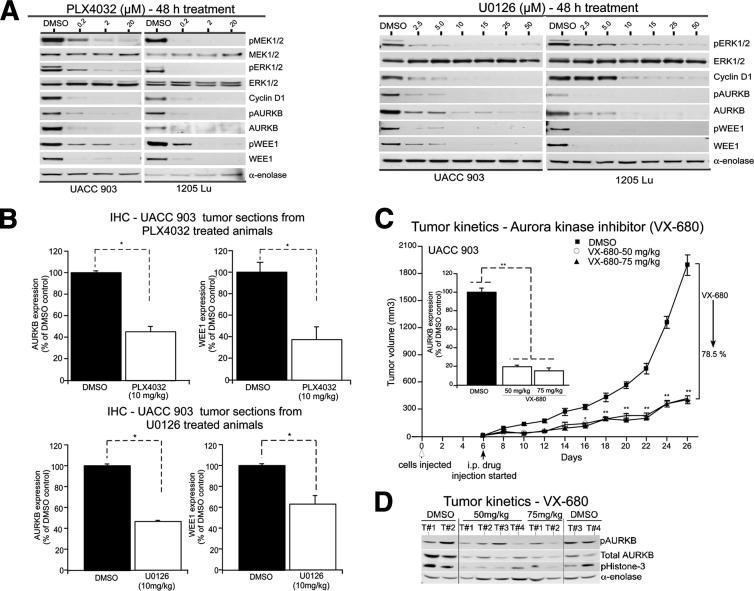
Figure 6.
AURKB or WEE1 serve as biomarkers of the activity of pharmacological agents targeting the MAP kinase pathway. A: UACC 903 or 1205 Lu cells were treated with 0.2 to 20 μmol/L vemurafenib, a B-RafV600E inhibitor (left panel), or 2.5 to 50 μmol/L U0126, an MEK1/2 inhibitor (right panel), for 48 hours. Protein lysates were collected and analyzed for MEK1/2, ERK1/2, cyclin D1, and AURKB or WEE1 expression by using Western blot analysis. α-Enolase served as a control for equal protein loading. A representative blot from two to three independent experiments is shown. B: Nude mice were injected with 5 × 106 UACC 903 melanoma cells, and 7 days later, vemurafenib or UO126 treatment at a dose of 10 mg/kg body weight was initiated, once per day for 6 days. Tumors were removed after last treatment and divided into sections for IHC staining of AURKB or WEE1. The expression of AURKB (top left panel) or WEE1 (top right panel) was quantified in tumor sections from vemurafenib-treated animals. Similarly, the expression of AURKB (bottom left panel) or WEE1 (bottom right panel) was quantified in tumor sections from U0126-treated animals compared with DMSO-treated controls and plotted. Data were obtained from three to four tumors, with four to five fields per tumor analyzed. C: A total of 1 × 106 UACC 903 melanoma cells were injected s.c. into the right and left flanks of nude mice, and 6 days later, they were treated daily i.p. with a dose of 50 or 75 mg/kg body weight of VX-680. Developing tumors were measured on alternate days up to day 26 and compared with controls treated with DMSO. Each point represents data obtained from five nude mice (two tumors on each side). Tumor sections were immunostained for AURKB and compared with DMSO control–treated tumors (inset). Data represent three to four tumors, with four to five fields per tumor analyzed. D: Protein lysates were analyzed by using Western blot analysis for AURKB, pAURKB, and pHistone-3. α-Enolase served as a control for equal protein loading. Data represent means ± SEM. *P < 0.01 (t-test) (B); *P < 0.01, **P < 0.001, analysis of variance, followed by a post hoc test (C).