Figure 4.
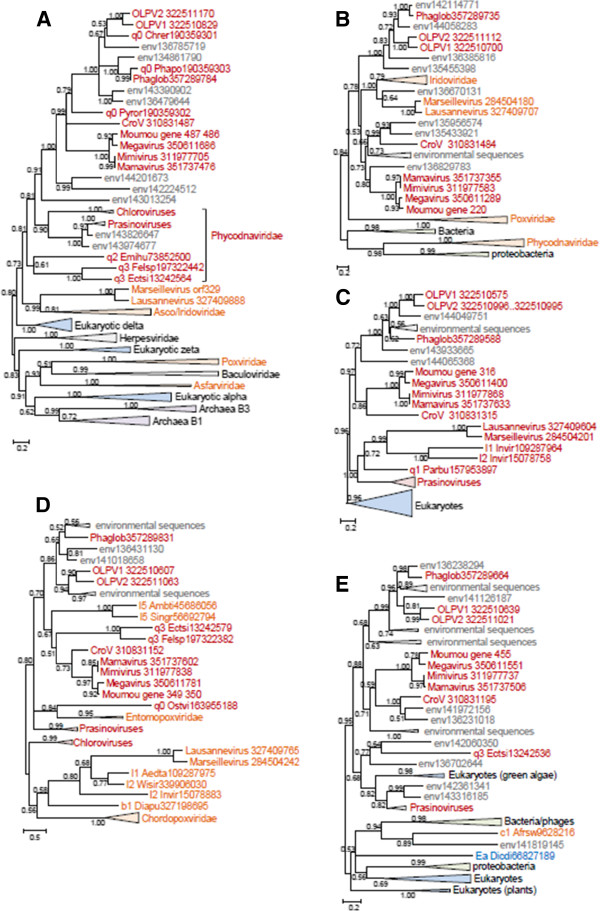
Maximum-Likelihood trees of ancestral NCLDV genes involved in DNA replication, recombination, and repair. A, DNA polymerase B, D5 primase-helicase. C, DNA topoisomerase II. D, Holliday junction (RuvC) resolvase. E, YqaJ-like recombinase. Branches with bootstrap support less than 0.5 were collapsed. For each sequence, the species name abbreviation and the gene identification numbers are indicated; env stands for “marine metagenome.” Species abbreviations: CroV, C.roenbergensis virus; Moumou, Moumouvirus; OLPV1, Organic Lake phycodnavirus 1; OLPV2, Organic Lake phycodnavirus 2; Phaglob, P. globosa virus; Aedta, Invertebrate iridescent virus 3; Afrsw, African swine fever virus; Ambti, Ambystomatigrinum virus; Chrer, Chrysochromulina ericina virus; Diapu, Diadromuspulchellus ascovirus 4a; Dicdi, Dictyostelium discoideum AX4; l1_Invir, Invertebrate iridescent virus 3; l2_Invir, Invertebrate iridescent virus 6; Ostvi, Ostreococcus virus OsV5; Parbu, Paramecium bursaria Chlorella virus AR158; Phapo, Phaeocystis pouchetii virus; Pyror, Pyramimonas orientalis virus; Singr, Singapore grouper iridovirus; Wisir, Wiseana iridescent virus; Ectsi, Ectocarpus siliculosus virus 1; Emihu, Emiliania huxleyi virus 86; Felsp, Feldmannia sp virus. Taxa abbreviations: Ea, Amoebozoa; b1, Ascovirus; c1, Asfarviridae; l1, Chloriridovirus; l2, Iridovirus; l5, Ranavirus; q0, unclassified Phycodnaviridae; q1, Chlorovirus; q2, Coccolithovirus; q3, Phaeovirus.
