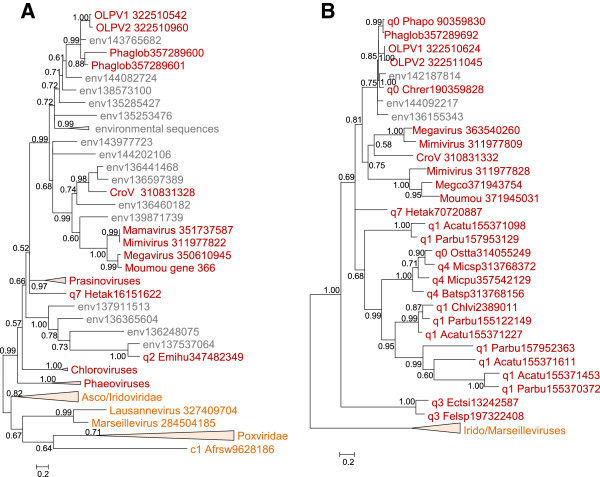
Figure 7.
Maximum-Likelihood trees of two genes involved in virion structure and morphogenesis. A, A32 virion packaging ATPase. B, Major capsid protein. Branches with bootstrap support less than 0.5 were collapsed. For each sequence, the species name abbreviation and the gene identification numbers are indicated; env stands for “marine metagenome.” Species abbreviations: Acatu, Acanthocystis turfacea Chlorella virus 1; Afrsw, African swine fever virus; Batsp, Bathycoccus sp. RCC1105 virus BpV1; Chlvi, Chlorella virus; Chrer, Chrysochromulina ericina virus; CroV , C. roenbergensis virus; Ectsi, Ectocarpus siliculosus virus 1; Emihu, Emiliania huxleyi virus 86; Felsp, Feldmannia species virus; Hetak, Heterosigma akashiwo virus 01; Megco, Megavirus courdo7; Micpu, Micromonas pusilla virus SP1; Micsp, Micromonas sp. RCC1109 virus MpV1; Moumou, Moumouvirus; OLPV1, Organic Lake phycodnavirus 1; OLPV2, Organic Lake phycodnavirus 2; Ostta, Ostreococcus tauri virus 2; Parbu, Paramecium bursaria Chlorella virus AR158; Phaglob, P. globosa virus; Phapo, Phaeocystis pouchetii virus. Taxa abbreviations: c1, Asfarviridae; q0, unclassified Phycodnaviridae; q1, Chlorovirus; q2, Coccolithovirus; q3, Phaeovirus; q4, Prasinovirus; q7, Raphidovirus.