Abstract
Background
Hydroxycinnamates (HCs) are mainly produced in plants. Caffeic acid (CA), p-coumaric acid (PA), ferulic acid (FA) and sinapic acid (SA) are members of the HC family. The consumption of HC by human might prevent cardiovascular disease and some types of cancer. The solubility of HCs is increased through thioester conjugation to various compounds such as quinic acid, shikimic acid, malic acid, anthranilic acid, and glycerol. Although hydroxycinnamate conjugates can be obtained from diverse plant sources such as coffee, tomato, potato, apple, and sweet potato, some parts of the world have limited availability to these compounds. Thus, there is growing interest in producing HC conjugates as nutraceutical supplements.
Results
Hydroxycinnamoyl transferases (HCTs) including hydroxycinnamate-CoA shikimate transferase (HST) and hydroxycinnamate-CoA quinate transferase (HQT) were co-expressed with 4-coumarateCoA:ligase (4CL) in Escherichia coli cultured in media supplemented with HCs. Two hydroxycinnamoyl conjugates, p-coumaroyl shikimates and chlorogenic acid, were thereby synthesized. Total 29.1 mg/L of four different p-coumaroyl shikimates (3-p-coumaroyl shikimate, 4-p-coumaroyl shikimate, 3,4-di-p-coumaroyl shikimate, 3,5-di-p-coumaroyl shikimate, and 4,5-di-p-coumaroyl shikimate) was obtained and 16 mg/L of chlorogenic acid was synthesized in the wild type E. coli strain. To increase the concentration of endogenous acceptor substrates such as shikimate and quinate, the shikimate pathway in E. coli was engineered. A E. coli aroL and aroK gene were mutated and the resulting mutants were used for the production of p-coumaroyl shikimate. An E. coli aroD mutant was used for the production of chlorogenic acid. We also optimized the vector and cell concentration optimization.
Conclusions
To produce p-coumaroyl-shikimates and chlorogenic acid in E. coli, several E. coli mutants (an aroD mutant for chlorogenic acid production; an aroL, aroK, and aroKL mutant for p-coumaroyl-shikimates production) were made and each mutant was tested using an optimized construct. Using this strategy, we produced 235 mg/L of p-coumaroyl-shikimates and 450 mg/L of chlorogenic acid.
Keywords: Chlorogenic acid, Hydroxycinnamic acid, Hydroxycinnamate-CoA quinate transferase, Hydroxycinnamate-CoA shikimate transferase
Background
Hydroxycinnamates (HCs) such as caffeic acid (CA), p-coumaric acid (PA), ferulic acid (FA) and sinapic acid (SA) are mainly produced in plants. Because HCs are anti-oxidants that can scavenge free radicals, it has been suggested that consumption of HCs might prevent cardiovascular disease and some types of cancer. In addition, several biological functions of HCs have been described, such as protection against side effects of chemotheraphy and anti-osteoclast activity [1].
In plants, HCs are usually conjugated with other compounds such as quinic acid, shikimic acid, malic acid, anthranilic acid, and glycerol [2]. Chlorogenic acid, the conjugate of CA and quinate, is abundant in coffee, fruits, and vegetables, and is the primary source of CA in the human diet [3]. For regular consumers of coffee in Western countries, the major dietary phenolics are chlorogenic acids [4].
The solubility of HCs is increased through thioester conjugation to various compounds. Although there is growing evidence for their beneficial health effects, the fruits and vegetables that contain high levels of HC conjugates are of limited availability in some parts of the world. Thus, there is growing interest in producing HC conjugates as nutraceutical supplements. The primary strategy for obtaining diverse phytochemicals is by extracting them from plants. However, this approach faces several obstacles such as a limited availability of plant materials and high costs of extraction and purification. Alternatively, they can be produced in microorganisms. Phytochemicals such as flavonoids and stilbenes have been produced using microorganisms such as Escherichia coli and Saccharomyces cerevisiae[5-7].
Biosynthesis of HC conjugates is mediated by hydroxycinnamoyl transferases (HCTs), which use the coenzyme A thioester of HCs as a donor and various compounds such as quinate, and shikimate as HC acceptors. The formation of coenzyme A thioester with HC is catalyzed by 4-coumaric acid:CoA ligase (4-CL) [8]. Thus, genes encoding 4-CL and HCT, are essential for producing HC conjugates from HC. HCTs from several plants have been characterized [8-12]. HCTs show specificity for not only the acyl group donor but also the acyl group acceptor. When the acyl group acceptors are shikimate and quinate, the resulting conjugates are p-coumaroyl-O-shikimate and chlorogenic acid, respectively [13,14].
E. coli is a good system for producing plant secondary metabolites including phytochemicals produced through the phenylpropanoid pathway [15]. We attempted to produce HC conjugates using E. coli. Shikimate and 3-dehydroquinate, which are acyl group acceptors for hydroxycinnamate-CoA shikimate transferases (HST) and hydroxycinnamate-CoA quinate transferases (HQT), respectively, can be synthesized through the shikimate pathway of E. coli[16,17]. Thus, it should be possible to alter the shikimate pathway to increase the concentrations of shikimate and quinate. However, the acyl donor (PA or CA) is a unique product of plants [18]. In order to produce HCs in E. coli, coexpression of several genes in E. coli is necessary, which leads to metabolic load. Therefore addition of exogenous HCs may circumvent this problem. In this report, we introduced either HST and 4CL (for the production of CA-shikimate), or HQT and 4CL (for the production of chlorogenic acid) into E. coli mutants that accumulate either shikimate or 3-hydroquinate. When the engineered cells were fed exogenous HCs, a high yield of HC-shikimate and HC-quinate were obtained.
Results
Construction and selection of the optimum expression vector for the synthesis of HC-shikimate
HC-shikimate was synthesized from HC-CoA and shikimate using HST (Figure 1). Two genes, HST from Nicotiana tabacum (NtHST) and 4CL from Oryza sativa (Os4CL), were subcloned into an E. coli expression vector. Os4CL converts the HCs into the corresponding HC-CoAs. Os4CL has a better catalytic efficiency than other 4CLs [19]. NtHST uses HC-CoAs such as p-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA as acyl donors and shikimate as an acyl acceptor to produce HC-shikimate. NtHST was the first characterized HST and only a few number of HST has been biochemically characterized. NtHST also uses quinate as an acyl acceptor [9]. Both genes (NtHST and Os4CL) were subcloned into three different E. coli expression vectors, each of which has a different copy number in E. coli. Each construct (pA-NtHST-Os4CL, pC-NtHST-Os4CL, or pE-NtHST-Os4CL; see Table 1) was transformed into E. coli BL21(DE3) cells, and tested for its ability to produce HC-shikimate when incubated with exogenous PA. As a control, the empty vector was transformed into E. coli. HCs including PA can enter into E. coli via the 4-hydroxyphenylacetate permease (HpaX transporter) [20] and small phenolic compounds can be exported from E. coli into the culture medium. The transformant harboring pC-NtHST-Os4CL produced the highest concentration of HC-shikimate derivatives (29.1 mg/L), whereas the titers of HC-shikimate derivatives produced by the transformants harboring pA-NtHST-Os4CL or pE-NtHST-Os4CL were 24.4 and 27.4 mg/L, respectively. However, only PA was observed in the transformant harboring the empty vector. According to ANOVA test, a significant difference in PA-shikimates production at P=0.01 was shown between pC-NtHST-Os4CL and pA-NtHST-Os4CL or between pE-NtHST-Os4CL and pA-NtHST-Os4CL, but not between pC-NtHST-Os4CL and pE-NtHST-Os4CL. The construct present in the pCDF vector (pC-NtHST-Os4CL) was used in further studies because this construct has a lower copy number than pE-NtHST-Os4CL. Therefore, pC-NtHST-Os4CL is expected to impose a lower metabolic load on E. coli.
Figure 1.
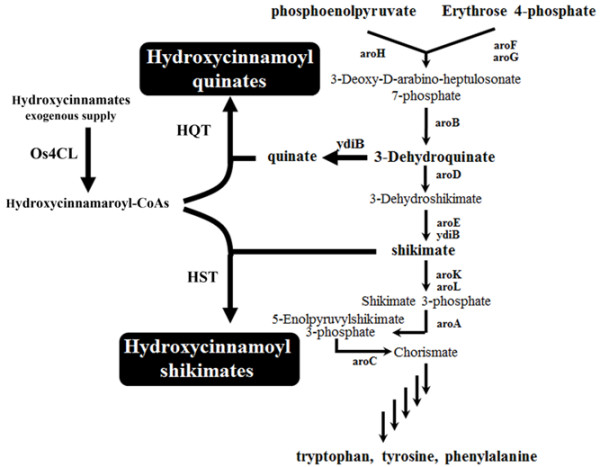
Schematic diagram showing the shikimate pathway of Escherichia coli and production of hydroxycinnamoyl quinate and hydroxycinnamoyl shikimate in E. coli.
Table 1.
Plasmids, Escherichia coli strains, and primers used in this study
| Plasmids or E. coli strains or Primers | Relevant properties or genetic marker | Source or reference |
|---|---|---|
| Plasmids | ||
| pCYCDuet |
P15A ori, Cmr |
Novagen |
| pCDFDuet |
CloDE13 ori, Strr |
Novagen |
| pETDuet |
f1 ori, Ampr |
Novagen |
| pA-NtHST-Os4CL |
pACYCDDuet carrying NtHST from N. tobacco and 4CL from O. sativa |
This study |
| pC-NtHST-Os4CL |
pCDFDuet carrying NtHST from N. tobacco and 4CL from O. sativa |
This study |
| pE-NtHST-Os4CL |
pETDuet carrying NtHST from N. tobacco and 4CL from O. sativa |
This study |
| pC-NtHQT-Os4CL |
pCDFDuet carrying NtHQT from N. tobacco and 4CL from O. sativa |
This study |
| pA-EcydiB |
pACYC carrying ydiB from E. coli |
This study |
| Strains |
|
|
| BL21 (DE3) |
F-ompT hsdSB(rB- mB-) gal dcm lon (DE3) |
Novagen |
| B-100 |
BaroD carrying pCDF-Duet |
This study |
| B-101 |
BaroD carrying pC-NtHQT-Os4CL |
This study |
| B-102 |
BaroD carrying pC-NtHQT-Os4CL and pA-EcydiB |
This study |
| BaroD |
BL21(DE3) ΔaroD::FRT-kanR-FRT |
This study |
| BaroK |
BL21(DE3) ΔaroK::FRT-kanR-FRT |
This study |
| BaroL |
BL21(DE3) ΔaroL::FRT-kanR-FRT |
This study |
| BaroKL |
BL21(DE3) ΔaroK::FRT ΔaroL::FRT-kanR-FRT |
This study |
| Primers |
|
|
| NtHST-F1 |
AACATATGAAGATCGAAGTGAAAGAAT (NdeI site is underlined) |
|
| NtHST-R2 |
AACTCGAGTCAAAAGTCATACAAGAACTTC (XhoI site is underlined) |
|
| NtHQT-F |
AAGATATCCATGGGAAGTGAAAAAATGATGA (EcoRV site is underlined) |
|
| NtHQT-R |
AAGGTACCTCAAAATTCATACAAATACTT (KpnI site is underlined) |
|
| ydiB-F |
ATGAATTCGATGGATGTTACCGCAAAATAC (EcoRI site is underlined) |
|
| ydiB-R |
CATGCGGCCGCTCAGGCACCGAACCCCATG (NotI site is underlined) |
|
| aroK-F |
gctgtcttttttacgctaatcttacccggtgatttatcgccagagcggtgaattaaccctcactaaagggcg |
|
| aroK-R |
cccgcagacgagtgtatataaagccagaattagttgctttccagcatgtgtaatacgactcactatagggctc |
|
| aroL-F |
atgacaccggctttcgccgcattgcgacctattggggaaaacccacgatgaattaaccctcactaaagggcg |
|
| aroL-R |
gatgaacgttaagtataggcgctcgaaaatcaacaattgatcgtctgtgctaatacgactcactatagggctc |
|
| aroD-F |
tggggttcggtgcctgacaggctgaccgcgtgcagaaagggtaaaaaatgaattaaccctcactaaagggcg |
|
| aroD-R |
gggaggatattcccgccgaaatattattgcttatgcctgatgtaaaatagttaatacgactcactatagggctc |
|
| aroK-check-F |
cgctgcctgcgttccatgat |
|
| aroL-check-F |
cgcggagctggagaagtggt |
|
| aroK or aroL check-R |
taatacgactcactatagggctc |
|
| aroD-check-F |
ggcaaggggctgaacagttc |
|
| aroD-check-R | gggaggatattcccgccgaa | |
1F means forward primer.
2R means reverse primer.
Although each vector exhibited varying biotransformation efficiencies, they all had the same reaction profiles. As shown in Figure 2, four new peaks appeared (P1 - P4). The molecular mass of P1 was 320.2 Da, indicating that one molecule of PA was attached to shikimate probably at the 5'-hydroxyl group of shikimate according to enzymatic studies [9,10]. In contrast, the molecular mass of the other three reaction products (P2 - P4) was 466.2 Da. Because the molecular mass of PA is 164 Da and that of shikimate is 174 Da, the three reaction products must be conjugates of two PA molecules and one shikimate molecule. p-Coumaroyl shikimates are not commercially available and the structures of the reaction products was determined using nuclear magnetic resonance spectroscopy (NMR). By the help of the HMBC and HMQC experiments (Additional file 1: Figure S1, S2), 14 13C peaks were determined, so that P1 was considered to be a conjugate of PA and shikimate. Even this structure contains 16 carbons, two set symmetric carbons of PA provide only two carbon peaks. Based on the HMBC spectrum (Additional file 1: Figure S1), the carboxyl group of PA was connected to the hydroxyl group of shikimate. Three isomers, the carboxyl group of PA connected to 3"-OH or 4"-OH or 5"-OH of shikimate, were possible. In the current NMR experiments, we could not determine the connected position. The name of the product P1 is (E)-3,4-dihydroxy-5-(3-(4-hydroxyphenyl)acryloyloxy)cyclohex-1-enecarboxylic acid where the bold number 5 could be switched with 3 or 4.
Figure 2.
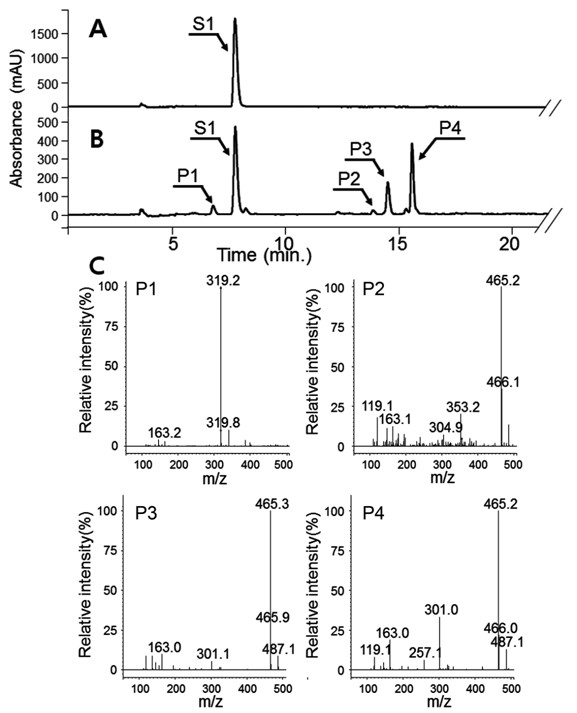
High performance liquid chromatography analysis of biotransformation of p-coumaric acid in BL21 (DE3) harboring Os4CL and NtHST. A, p-coumaric acid (S1); B, reaction products of p-coumaric acid (P1 - P4); C, mass spectrometry (MS) data for the four reaction products. The mass spectrometer was operated in negative mode. The molecular masses of p-coumaric acid and shikimic acid are 164 Da and 174 Da, respectively.
Based on the molecular masses of P2, P3, and P4 (Figure 2C), they contained two PAs and one shikimate. Two PAs can be connected to 3"-OH or 4"-OH or 5"-OH of SA, so that there can be three isomers, i.e. 3"-OH/4"-OH, 3"-OH/5"-OH, or 4"-OH/5"-OH. We could not determine which produce was which isomer. By comparison to a previous report [21], P3 and P4 are likely to be 3,5-di-p-coumaroyl shikimate and 4,5-di-p-coumaroyl shikimate, respectively. The remaining peak, P2 is therefore likely to be 3,4-di-p-coumaroyl shikimate.
Using E. coli harboring pC-NtHST-Os4CL, we determined the best acyl donor among CA, PA, and FA. In a previous study, recombinant NtHST protein most efficiently used caffeoyl-CoA as an acyl donor [9]. In this study, 1 mM of each HC was added to the same number of cells, and biotransformation was performed for 6 h. PA was the most effective acyl donor, producing 23.9 mg/L of p-coumaroyl shikimate. The amounts of feruloyl shikimate and caffeoyl shikimate produced were 3.8 mg/L, and 3.1 mg/L, respectively. Taken together, these results indicate that the highest amount of HC-shikimate was obtained with pC-NtHST-Os4CL as the construct and PA as the substrate.
Engineering E. coli to increase production of PA-shikimate conjugates
Because PA was added to the E. coli culture as an acyl donor, it was not a limiting factor for the production of PA-shikimate conjugates. However, E. coli uses endogenous shikimate as an acyl group acceptor. Thus, limiting shikimate availability would cause a bottleneck in the production of PA-shikimate conjugate. Previous studies showed that deletion of aroK and/or aroL in E. coli results in increased shikimate accumulation [22-24]. We created an E. coli aroL deletion mutant (BaroL), an aroK deletion mutant (BaroK) and an aroK/aroL double mutant (BaroKL) by gene replacement. pC-NtHST-Os4CL was transformed into each mutant and into wild type. BaroL showed the highest productivity (approximately 235 mg/L), which is approximately 15-fold higher than BL21 (16 mg/L). BaroKL and BaroK produced 228 mg/L and 171 mg/L of PA-shikimates, respectively (Figure 3). According to ANOVA test, PA-shikimates production was significantly different at P=0.01 level among BL21, BaroK, and BaroL. However, there was no difference between BaroL and BaroKL at this level of significance.
Figure 3.
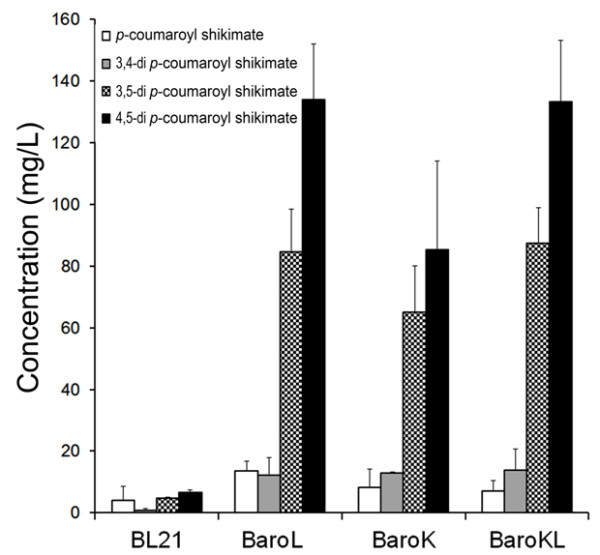
Production of hydroxycinnamic shikimates in different E. coli strains. Product 1 contains one molecule of hydroxycinnamate attached to shikimate, while products 2–4 contain two molecules of hydroxycinnamate attached to shikimate. Error bars indicate mean values ± SD from three independent experiments. Product 1 is 3 or 4-p-coumaroyl shikimate; The structures of product 2, product 3, and product 4 are likely to be 3,4-di-p-coumaroyl 3,5-di-p-coumaroyl shikimate, and 4,5-di-p-coumaroyl shikimate, respectively.
Next, production of PA-shikimate was optimized using BaroL harboring pC-NtHST-Os4CL. The optimum cell concentration was determined by varying the cell concentration from OD600 1 to 5 in the presence of 1 mM PA. Production of PA-shikimate peaked at an OD600 of 3, at which cell concentration the yield of PA-shikimate was approximately 235 mg/L. At OD600 = 1, or 2, the yield was approximately 94 mg/L, and 169 mg/L, respectively. Above OD600 = 3, the production of PA-shikimate decreased and was approximately 188 or 103.7 mg/L at OD600 = 4 or 5, respectively.
Using BaroL harboring pC-NtHST-Os4CL at a cell concentration of OD600 3, we monitored the production of HC-shikimate over 12 h. The maximum amount of di-p-coumaroyl shikimates was produced at 8 h (134 mg/L 3,5-di-p-coumaroyl shikimate, 70 mg/L 4,5-di-p-coumaroyl shikimate, 8 mg/L of 3,4-di-p-coumaroyl shikimate, and 23 mg/L of 5-p-coumaroyl shikimate). Therefore, a total of 235 mg/L of PA-shikimates was produced. At 8 h, the amount of 4,5-di-p-coumaroyl shikimate and 3,5-di-p-coumaroyl shikimate was reduced, but the amount of 5-p-coumaroyl shikimate reached a maximum (Figure 4). It seems that 5-p-coumaroyl shikimate was immediately converted to either 3,5-di-p-coumaroyl shikimate or 4,5-di-p-coumaroyl shikimate before 8 h.
Figure 4.
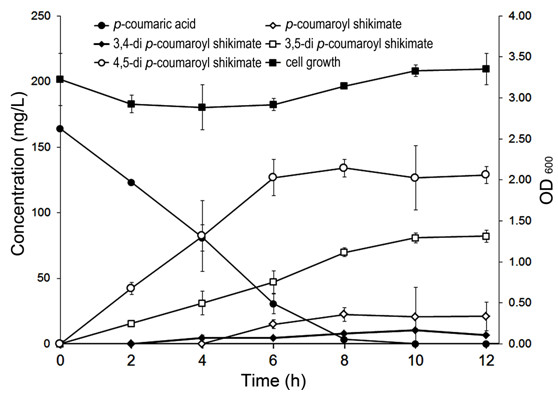
Production of hydroxycinnamic shikimates by biotransformation using BaroL harboring pC-NtHST-Os4CL. Error bars indicate mean values ± SD from three independent experiments.
Production of HC-quinate in E. coli
We used an acyltransferase that is specific for quinate to produce HC-quinate. Hydroxycinnamoyl-CoA quinate transferase from N. tobacco (NtHQT) was shown to be more specific for quinate than for shikimate [9,25]. Os4CL mediates formation of coenzyme A thioester with HC. To produce HC-quinate in E. coli, two genes, NtHQT and Os4CL, were subcloned into an E. coli expression vector. The resulting construct, pC-NTHQT-Os4CL was transformed into BL21(DE3), and the transformant was used for biotransformation of CA. However, no reaction product was detectable. The amount of quinate or dehydroquinate in E. coli may not be high enough to drive the production of chlorogenic acid. Therefore, we created strain BaroD, which should accumulate higher levels of dehydroquinate and/or quinate, in order to overcome this potential limitation. The strain BaroD was used as a host for pC-NTHQT-Os4CL and the resulting transformant was called B-101 (Table 1). As control, empty pCDFDuet was transformed into the strain BaroD and this transformant was called B-100 (Table 1). Each transformant was used for biotransformation of CA. The culture medium of B100 became a brown color after 8 h. Analysis of the reaction mixture using HPLC after 8 h incubation showed a trace level of caffeic acid. In addition, no detectable new product was observed. The decrease of caffeic acid during biotransformation is associated with the formation of O-quinone and its polyaromatic derivatives [26]. However, analysis of the biotransformation mixture from strain B-101 showed a new peak (Figure 5C) and the color of the culture medium did not change. Once caffeic acid is converted into another compound, it cannot polymerize and therefore the color of the culture medium does not change. The molecular mass of the product was 352 Da, which was 2 Da less than the predicted molecular mass of the thioester of quinate and CA (Figure 5C and E). Based on the molecular weight of the reaction product and the known intermediates of the E. coli shikimate pathway, it is like that 3-dehydroquinate and not quinate was attached to CA. NtHQT utilized 3-dehydroquinate as an acceptor and caffeoyl-CoA as a donor to make caffeoyl-3-dehydroquinate. Expression of ydiB from E. coli, which converts dehydroquinate into quinate, also increased the production of HC ester yield because NtHQT was likely to have higher affinity for quinate than for dehydroquinate as acyl group acceptor.
Figure 5.
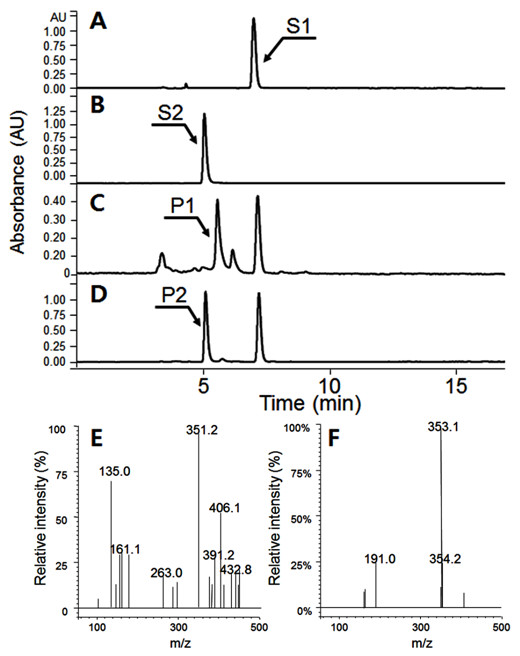
Production of chlorogenic acid in E. coli. A, caffeic acid (S1); B, chlorogenic acid (S2); C, reaction product of caffeic acid obtained from strain B-101; D, reaction product of caffeic acid obtained from strain B-102; E, MS/MS profile of P1; F, MS/MS profile of P2.
The protein product of ydiB converts 3-dehydroquinate into quinate [27]. ydiB was overexpressed in E. coli strain B-101 to make strain B-102. Biotransformation of CA by strain B-102 resulted in a new product with an identical HPLC retention time and molecular mass (354 Da) as chlorogenic acid (Figure 5D, F). The MS/MS spectrum of the reaction product matched authentic chlorogenic acid. This indicates that ydiB converts 3-dehydroquinate into quinate, which was then utilized for the production of chlorogenic acid.
PA and FA were also tested as acyl-group donors. CA was the best acyl-group donor followed by PA and FA. After 24 h, the amounts of caffeoyl-quinate, p-coumaroyl-quinate, and feruloyl-quinate were 450 mg/L, 323.7 mg/L, and 216 mg/L, respectively.
The production of chlorogenic acid was monitored further in strain B-102. After induction, 1.1, 1.3, 1.5, 1.7, or 2.0 mM CA was added to the culture. After 24 h, almost all of the CA was converted into chlorogenic acid in the reaction mixtures supplied with 1.1, 1.3, or 1.5 mM of CA. However, at a concentration of 1.7 or 2.0 mM, some CA remained in the mixture, and even after a longer incubation time, the remaining caffeic acid was not converted into chlorogenic acid. However, the color of the mixture became dark brown due to the oxidation/polymerization of CA. Therefore, we monitored chlorogenic acid production at a CA concentration of 1.5 mM. As shown in Figure 6, production of chlorogenic acid continued to increase until 24 h, while the CA content decreased. Approximately 450 mg/L of chlorogenic acid was produced at 24 h.
Figure 6.
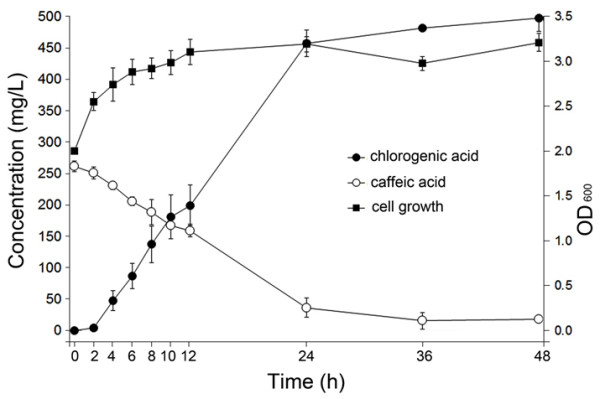
Production of chlorogenic acid using the E. coli strain B-106.
Discussion
Anthocyanins, flavonoids, stilbene, and other compounds of plant origins have been biologically synthesized using engineered E. coli strains [7,15]. Most of these efforts involved introduction of new genes with high activity into E. coli. In addition, cofactor supplementation and reducing equivalents have been enhanced to produce natural compounds using engineered E. coli[6,28]. The goal of this study was to produce PA-shikimate and chlorogenic acid in E. coli. We also engineered the shikimate pathway of E. coli to accumulate the acyl group acceptors, shikimate and quinate, which led to the increased production of PA-shikimates and chlorogenic acid.
During biotransformation, two PA molecules are attached to shikimate, whereas only one molecule of CA is attached to quinate. In plants, one molecule of CA is bound to one molecule of shikimate. However, diverse HC-quinate conjugates, including those in which two or three identical HCs are bound to quinate, and two or three different HCs are bound to quinate, were found in coffee (Coffea robusta) and sweet potato (Ipomonea batatas) [21,29]. In these plants, the mono-esters are present as major components whereas di- and tri-esters are present as minor components. NtHST and NtHQT may have different enzymatic properties from HCTs from coffee or sweet potato because di-p-coumaroyl skimates and cholorgenic acid were synthesized using NtHST and NtHQT. One possible scenario is that the substrate binding pocket of NtHQT may be narrower than that of NtHST. Thus, HC-quinate may not fit into the substrate binding pocket of NtHQT for the second round of acyl transfer reaction. Longer incubation of the recombinant NtHST enzyme with PA-CoA and shikimate resulted in the production of di-PA-shikimate (data not shown). The production of di-PA-shikimate was also observed with HST from Populus euramericana[13]. Using the properties of NtHST, diverse HC-shikimate conjugates could possibly be obtained and feeding of different concentrations of various HCs into the E. coli culture medium may result in production of various forms of HC-shikimate conjugates. Any biological activity including anti-oxidant activity of di-PA-shikimate compared to PA-shikimate has not been tested. However, 8-O-4-diferulic acid showed better antioxidant activity than ferulic acid [30].
p-Coumaric acid can be synthesized from tyrosine by the action of tyrosine ammonia lyase (TAL). Therefore, synthesis of p-coumaroyl-shikimate or chlorogenic acid from glucose might be possible if a gene econding TAL were expressed into E. coli harboring NtHST and Os4CL or into E. coli harboring NtHQT, Os4CL, ydiB, and Sam5 (Sam5 converts p-coumaric acid into caffeic acid [31]), respectively. However, it is expected that only a tiny amount of reaction product would be produced from glucose in a wild type strain because quinate and shikimate are not accumulated in the wild type E. coli. Use of strain BaroD or BaroL is not also feasible because tyrosine is not synthesized in these strains. Thus, supplementation with either caffeic acid or tyrosine is needed to synthesize p-coumaroyl-shikimate or chlorogenic acid in E. coli.
Wild type E. coli expressing NtHQT and Os4CL did not produce detectable amounts of chlorogenic acid or caffeoyl-dehydroquinate after CA supplementation. In the wild type, dehydroquinate may have been rapidly converted into another downstream compound of the shikimate pathway instead of accumulating. Moreover, the expression level of ydiB, which converts dehydroquinate into quinate, was low. Only a small amount of quinate or dehydroquinate will be expected to be present in the cell in the absence of overexpression of ydiB[32]. ydiB and aroE mediate the conversion reaction not only from dehydroquinate to quinate but also from dehydroshikimate to shikimate. However, aroE prefers dehydroshikimate to dehydroquinate [22], whereas ydiB has nearly the same catalytic efficiency for dehydroshikimate and dehydroquinate [27,33]. Recent studies have shown that overexpression of ydiB does not increase shikimate, while overexpression of aroE increases conversion of dehydroshikimate to shikimate [32]. Therefore, we overexpressed ydiB to produce chlorogenic acid. In addition, strains BaroK, BaroL, and BaroKL expressing pC-NtHQT-Os4CL that were supplemented with CA produced chlorogenic acid only when ydiB was overexpressed (data not shown). This indicated that these mutants accumulate shikimate pathway intermediates such as dehydroquinate, dehydroshikimate and shikimate. These intermediates are converted into quiniate by ydiB.
Conclusions
To produce the HC-conjugates in E. coli, several E. coli mutants including aroD, aroK, aroL, and aroK/L mutants were made and each mutant was tested using an optimized construct. For the production of HC-shikimates, an E. coli aroL mutant (B-aroL) was best and we produced 235 mg/L of HC-shikimates using B-aroL expressing NtHST and Os4CL, which is approximately 15-fold higher than wild type E. coli BL21 (16 mg/L). The wild type E. coli expressing NtHQT and Os4CL did not produce any detectable chlorogenic acid. By using E. coli aroD mutant (strain B-101), which accumulated 3-dehydroquinate, caffeoyl-3-dehydroquinate instead of chlorogenic acid, was synthesized. However, by expressing ydiB gene in strain B-101, which converted 3-dehyroquinate to quinate, chlorogenic acid production was dramatically increased up to 450 mg/L.
Materials and methods
Strains and reagents
The E. coli strains used in this study are listed in Table 1. E. coli BL21 (DE3) cells were used for recombinant protein production. E. coli DH5α cells were used for plasmid cloning. All restriction enzymes and T4 DNA ligase were purchased from Takara (Shiga, Japan). Polymerase chain reaction (PCR) amplification was performed using Hotstart Taq DNA polymerase (Qiagen, Hilden Germany). Reverse transcription was performed using Omniscript reverse transcriptase (Qiagen). E. coli was cultured in Luria-Bertani (LB) or M9 medium (plus 2% glucose) containing 50 μg/mL antibiotics, when necessary. E. coli expression vectors were purchased from Novagen (Madison, WI, USA).
Construction of E. coli expression vector
The genes for hydroxycinnamate-CoA shikimate transferase (NtHST) [9] and hydroxycinnamate-CoA quinate transferase (NtHQT) from tobacco (N. tabacum) [26] were cloned using reverse-transcription polymerase chain reaction (RT-PCR). Total RNA was isolated from the leaves of one-month-old tobacco using Plant Total RNA Isolation Kit (Qiagen), and cDNA was synthesized using Omniscript reverse transcriptase (Qiagen) and oligo dT as a primer. PCR was carried out using primers designed on the basis of the published sequences (GenBank accession AJ507825 for NtHST and AJ582651 for NtHQT). The primers were listed in Table 1. The Os4CL gene, which was cloned and characterized previously from rice [19], was subcloned into the BamHI/NotI sites of pACYCDuet, pCDFDuet, and pETDuet vectors (EMD Chemicals, Gibbstown, NJ, USA), and then the resulting NtHST PCR product was subcloned into a second cloning site, the NdeI/XhoI site of each vector. The NtHQT PCR product was subcloned into the EcoRV/KpnI site of pCDFDuet, which contains Os4CL at NdeI/XhoI site. The resulting constructs are listed in Table 1.
Deletion of the aroL and aroK genes in E. coli BL21(DE3) was accomplished using the Quick and Easy Conditional Knockout Kit (Gene Bridges, Heidelberg, Germany). Briefly, the aroL gene or the aroK gene of E. coli BL21 (DE3) was replaced by the ΔaroL FRT-PGK-gb2-neo-FRT cassette or the ΔaroK FRT-PGK-gb2-neo-FRT cassette, respectively [34]. Deletion mutants were selected in Luria-Bertani (LB) medium containing 50 μg/mL kanamycin. Deletion of aroL or aroK was confirmed using PCR. The strains deleted in aroL or aroK were named BaroL and BaroK, respectively (Table 1). ΔaroL and ΔaroK double mutant (strain BaroKL in Table 1) was constructed using strain BaroL. The kanamycin cassette was removed in ΔaroL using an FLP expression plasmid, which removes the kanamycin selection marker from the chromosome. Removal of the kanamycin cassette was confirmed by PCR. Using ΔaroL as a host, the aroK gene was replaced by the ΔaroK FRT-PGK-gb2-neo-FRT cassette. The primer sequences for the aroD, aroK, and aroL deletion are shown in Table 1.
Shikimate/quinate dehydrogenase gene, ydiB was cloned using E. coli BL21 (DE3) genomic DNA as a template. Primers were designed based on the published sequence (NC_000913.2) and the sequences of primers were listed in Table 1. The PCR product was digested with EcoRI/NotI and subcloned into the corresponding site of pCDF-Duet1. The resulting constructs were confirmed by sequencing.
Production of HC-shikimate conjugate in E. coli
Each construct was transformed into E. coli BL21 (DE3) strain or BaroL strain using electroporation with the BioRad MicroPulser Electroporation Apparatus (BioRad, Hercules, CA, USA). Overnight cultures of transformants were inoculated into LB medium containing 50 μg/mL of antibiotic at 37°C and cultured until the OD600 reached 0.8. Protein expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and growth was continued for another 20 h at 18°C. Cells were harvested and resuspended to obtain a cell concentration corresponding to OD600 of 3 in 2 mL of fresh M9 medium containing 2% glucose, 50 μg/ mL of antibiotics, 1 mM IPTG, and 1 mM PA in a test tube (14 mm × 145 mm) and then cultured at 30°C for 8 h. The supernatant was extracted twice using an equal volume of ethyl acetate, and the upper aqueous phase was dried using a Speed Vac at 30°C, dissolved in 60 μL of dimethylsulfoxide (DMSO), and analyzed using high-performance liquid chromatography (HPLC). Because p-coumaroyl shikimate is not commercially available, we used p-coumaric acid to generate a standard curve for quantitative analysis of the reaction products. The UV spectra of p-coumaric acid are distinguishable from those of p-coumaroyl shikimate but they are very similar and the absorption of the thioesters is usually shifted to longer wavelength by a few nm. This is subtle enough that it should be perfectly acceptable to use the free acids as quantitation standards for the thioesters.
To determine the maximum conversion rate using the optimized vector, protein expression was induced as described above. The cell concentration was adjusted to an OD600 of 3 in 25 mL of fresh M9 containing 2% glucose, 1 mM IPTG, and 50 μg/mL of antibiotics. The medium was supplemented with 1 mM PA. The reaction product (200 μL) was collected and extracted with ethyl acetate. The supernatant was dried and dissolved with 100 μl of dimethyl sulfoxide (DMSO). The sample was directly injected to HPLC for analysis. The mean and the standard error of the mean were calculated from triplicate experiments. Analysis of variance (ANOVA) was carried out using Tukey’s method with a significance level of P=0.01 using 2010 Microsoft Office Excel.
Production of chlorogenic acid in E. coli
The construct for the production of chlorogenic acid was transformed into E. coli BL21(DE3) or BaroD cells. Induction of each construct was performed as described above. To determine the optimal gene construct, 200 μM PA was added to the growth medium.
To measure the production of chlorogenic acid in BaroD cells harboring pC-EcycdiB and pC-NtHQT-Os4CL (Table 1), the cell concentration was adjusted to an OD600 of 2.0, and CA was added to the medium to a final concentration of 1.5 mM. Production of chlorogenic acid was periodically monitored. The biotransformation was stopped by boiling for 5 min and the biotransformation product was centrifuged for 15 min at 13000 × g to remove the cell debris and other components prior to HPLC analysis. The quantification of the product was carried out using a standard curve generated with authentic chlorogenic acid (Sigma, MO, USA).
Analysis of the metabolites
The metabolites were analyzed using a Varian HPLC equipped with a photo diode array (PDA) detector and a Varian C18 reversed-phase column (Varian, 4.60 × 250 mm, 3.5 μm particle size). The mobile phases consisted of 0.1% formic acid in water and acetonitrile. For chlorogenic acid, the program was: 20% acetonitrile at 0 min, 32% acetonitrile at 15 min, 90% acetonitrile at 17 min, 90% acetonitrile at 20 min, 20% acetonitrile at 21 min, and 20% acetonitrile at 26 min. To analyze hydroxycinnamoyl shikimate, the program was: 25% acetonitrile at 0 min, 40% acetonitrile at 10 min, 75% acetonitrile at 15 min, 90% acetonitrile at 22 min, 25% acetonitrile at 23 min, and 25% acetonitrile at 30 min. The flow rate was 1 mL/min, and the separation was monitored at 290 nm and 320 nm.
The molecular masses of the metabolites were determined using a Varian 500-MS ion trap spectrometer. Mass spectra were acquired simultaneously using an electrospray ionization source in negative ionization mode at 600 V. NMR spectrometry was done as described before [35].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JHA initiated and coordinated the project. BGK, JWD, HM, and JHA performed experiments, analyzed data and wrote the paper. All authors approved the final manuscript.
Supplementary Material
The HMBC spectrum of the product P1. Figure S2. The HMQC spectrum of the product P1.
Contributor Information
Bong-Gyu Kim, Email: dkimbk@hanmail.net.
Woo Dam Jung, Email: wjddneka@cyworld.com.
Hyejung Mok, Email: hjmok@konkuk.ac.kr.
Joong-Hoon Ahn, Email: jhahn@konkuk.ac.kr.
Acknowledgements
This work was supported by a grant from Systems and Synthetic Agro-Biotech Center through the Next-Generation BioGreen 21 Program (PJ00948301), Rural Development Administration, and partially funded by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012–0006686).
References
- El-Seedi HR, El-Seed AMA, Khalifa SAM, Görasson U, Bohlin L, Borg-Karlson A-K, Verpoorte R. Biosynthesis, natural sources, dietary intake, pharmacokinetic properties, and biological activities of hydroxycinnamic acids. J Agri Food Chem. 2012;60(44):10877–10895. doi: 10.1021/jf301807g. [DOI] [PubMed] [Google Scholar]
- Clifford MN. Chlorogenic acids and other cinnamates-nature, occurrence and dietary burden. J Sci Food Agric. 1999;79(3):362–372. doi: 10.1002/(SICI)1097-0010(19990301)79:3<362::AID-JSFA256>3.0.CO;2-D. [DOI] [Google Scholar]
- Herrmann K. Occurrence and content of hydroxycinnamic acid and hydoxybenzoic acid compounds in foods. Crit Rev Food Sci Nutr. 1989;28(4):315–347. doi: 10.1080/10408398909527504. [DOI] [PubMed] [Google Scholar]
- Crozier A, Jaganatha IB, Clifford MM. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26(8):1001–10043. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- Chemler JA, Koffas MAG. Metabolic engineering for plant natural product biosynthesis in microbes. Cur Opin Biotech. 2008;19(6):597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Chemler JA, Fowler ZL, Mchugh KP, Koffas MAG. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng. 2010;12(2):96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Fowler ZL, Koffas MAG. Biosynthesis and biotechnological production of flavanones: current state and perspectives. Appl Microbiol Biotechnol. 2009;83(5):799–808. doi: 10.1007/s00253-009-2039-z. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichershy E. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem. 2002;302(2):305–312. doi: 10.1006/abio.2001.5574. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem. 2003;278(1):95–103. doi: 10.1074/jbc.M209362200. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffoy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. Silencing of hydroxycinnamoyl-Coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 2004;16(6):1446–1465. doi: 10.1105/tpc.020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Kiri LT. Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Aca Sci USA. 2007;104(28):11856–11861. doi: 10.1073/pnas.0701428104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comino C, Lanteri S, Portis E, Acquadro A, Romani A, Hehn A, Larbat R, Bourgaud F. Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol. 2007;20(7):14. doi: 10.1186/1471-2229-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-G, Lee ET, Ahn J-H. Characterization of hydroxycinnamoyl-coenzyme A shikimate hydroxycinnamoyltransferase from Populus euramericana. J Kor Soc Appl Biol Chem. 2011;54(2):817–821. [Google Scholar]
- St Pierre B, De Luca V. In: Recent Advances in Phytochemistry Vol 34. Evolution of Metabolic Pathways. Romeo JT, Ibrahim R, Varin L, De Luca V, editor. Oxford: Elsevier Science Ltd; 2000. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism; pp. 285–315. [Google Scholar]
- Horinouchi S. Combinatorial biosynthesis of non-bacterial and unnatural flavonoids, stilbenoids and curcuminoids by microorganisms. J Antibiot. 2008;6(12):709–728. doi: 10.1038/ja.2008.85. [DOI] [PubMed] [Google Scholar]
- Ikeda M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl Microbiol Biotechnol. 2006;69(6):615–626. doi: 10.1007/s00253-005-0252-y. [DOI] [PubMed] [Google Scholar]
- Gosset G. Production of aromatic compounds in bacteria. Curr Opin Biotech. 2009;20(6):651–658. doi: 10.1016/j.copbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7(7):1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeeY J, Jeon Y, Lee JS, Kim B-G, Lee CH, Ahn J-H. Enzymatic synthesis of phenolic CoAs using 4-coumarate:coenzyme A ligase (4CL) from rice. Bull Kor Chem Soc. 2007;28(3):365–366. [Google Scholar]
- Diaz E, Ferrández A, Prieto MA, Garcia JL. Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev. 2001;65(4):523–569. doi: 10.1128/MMBR.65.4.523-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R, Patras MA, Eravuchira PJ, Kuhnert N. Profile and characterization of the chlorogenic acids in green robusta coffee beans by LC-MSn: Identification of seven new classes of compounds. J Agric Food Chem. 2010;58(15):8722–8737. doi: 10.1021/jf1014457. [DOI] [PubMed] [Google Scholar]
- Draths KM, Knop DR, Frost JW. Shikimic acid and quinic acid: replacing isolation from plant sources with recombinant microbial biocatalysis. J Am Chem Soc. 1999;121(7):1603–1604. doi: 10.1021/ja9830243. [DOI] [Google Scholar]
- Escalante A, Calderón R, Valdivia A, de Anda R, Hernánde G, Ramirez OT, Gosset G, Bolivar F. Metabolic engineering for the production of shikimic acid in an evolved Esherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system. Microbial Cell Fact. 2010;9:21–33. doi: 10.1186/1475-2859-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer M, Bongaerts J, Bovenberg R, Kremer S, Müller U, Orf S, Wubbolts M, Raeven L. Metabolic engineering for microbial production of shikimic acid. Met Eng. 2003;5(4):277–283. doi: 10.1016/j.ymben.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotech. 2004;22(4):746–754. doi: 10.1038/nbt966. [DOI] [PubMed] [Google Scholar]
- Zhang H, Stephanopoulos G. Engineering E. coli for caffeic acid biosynthesis from renewable sugar. Appl Microbiol Biotechnol. 2012. [DOI] [PubMed]
- Lindner HA, Nadeau G, Matte A, Michel G, Ménard R, Cygler M. Site-directed mutagenesis of the active site region in the quinate/shimate 5-dehydrogenase YdiB of Escherichia coli. J Biol Chem. 2005;280(8):7162–7169. doi: 10.1074/jbc.M412028200. [DOI] [PubMed] [Google Scholar]
- Sung SH, Kim BG, Ahn J-H. Optimization of rhamnetin production in Escherichia coli. J Microbio Biotech. 2011;21(8):854–857. doi: 10.4014/jmb.1104.04048. [DOI] [PubMed] [Google Scholar]
- Zheng W, Clifford MN. Profiling the chlorogenic acids of sweet potato (Ipomoea batatas) from China. Food Chem. 2010;106(1):147–152. [Google Scholar]
- Garcia-Conesa MT, Plumb GW, Waldron KW, Ralph J, Williamson G. Ferulic acid dehydrodimers from wheat bran: isolation, purification and antioxidant properties of 8-O-4-diferulic acid. Redox Rep. 1997;3(5–6):319–323. doi: 10.1080/13510002.1997.11747129. [DOI] [PubMed] [Google Scholar]
- Berner M, Krug D, Bihlmaier C, Vente A, Müller R, Bechthold A. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothix espanaensis. J Bact. 2006;188(7):2666–2673. doi: 10.1128/JB.188.7.2666-2673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juminaga D, Baidoo EE, Redding-Johanson AM, Batth TS, Burd H, Mukhopadhyay A, Petzold CJ, Keasling JD. Modular engineering of L-tyrosine production in Escherichia coli. Appl Environ Microbiol. 2012;78(1):89–98. doi: 10.1128/AEM.06017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G, Roszak AW, Sauvé V, Maclean J, Matte A, Coggins JR, Cygler M, Lapthorn AJ. Structure of shikimate dehydrogenase AroE and its paralog YdiB. J Biol Chem. 2003;278(21):19463–19472. doi: 10.1074/jbc.M300794200. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-A, Kim B-G, Lee WJ, Lim Y, Chong Y, Ahn J-H. Production of a novel quercetin glycoside through metabolic engineering of Escherichia coli. Appl Env Microbiol. 2012;78(12):4256–4262. doi: 10.1128/AEM.00275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The HMBC spectrum of the product P1. Figure S2. The HMQC spectrum of the product P1.


