Abstract
In metazoans with “open” mitosis, cells undergo structural changes involving the complete disassembly of the nuclear envelope (NE). In post-mitosis, the dividing cell faces the difficulty to reassemble NE structures in a highly regulated fashion around separated chromosomes. The de novo formation of nuclear pore complexes (NPCs), which are gateways between the cytoplasm and nucleoplasm across the nuclear membrane, is an archetype of macromolecular assembly and is therefore of special interest. The reformation of a functional NE further involves the reassembly and organization of other NE components, the nuclear membrane and NE proteins, around chromosomes in late mitosis.
Here, we discuss the function of NE components, such as lamins and INM proteins, in NE reformation and highlight recent results on coordination of NPC and NE assembly.
Keywords: nuclear envelope, nuclear pore complex, inner nuclear membrane, nuclear reassembly, open mitosis, lamin, nucleoporin
Introduction
An overview of the nuclear envelope (NE)
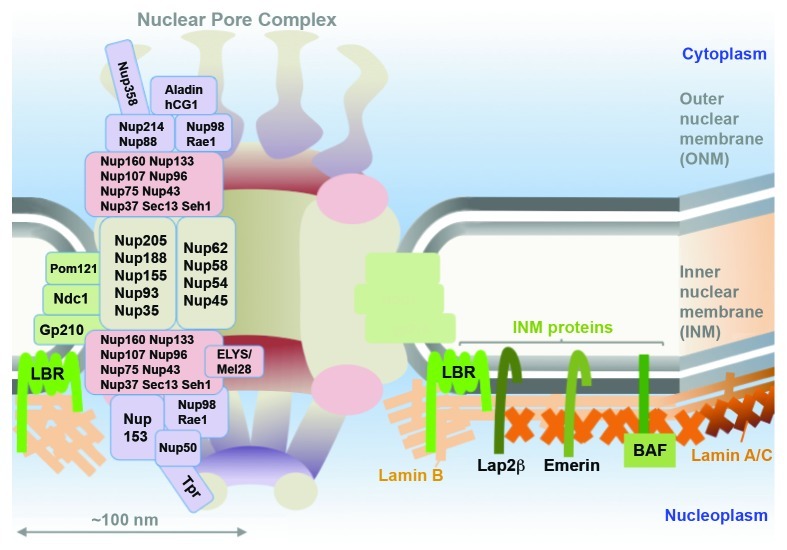
The nuclear envelope (NE) is the physical barrier between the cytoplasm and nucleoplasm. The NE is composed of two lipid bilayer membranes: the outer nuclear membrane (ONM), which is continuous with the endoplasmic reticulum (ER), and the inner nuclear membrane (INM), which contains a specific subset of more than 60 INM or NE transmembrane proteins.1 The ONM and the INM, which are ~20−40 nm apart,2 fuse at places where the nuclear pore complex (NPC) is embedded. NPCs provide an aqueous channel for molecular exchange between the cytoplasm and nucleoplasm. The NPC is a large structure (60−125 MDa) with octahedral symmetry assembled by multicopies of ~30 different proteins called nucleoporins or Nups.3,4
Small molecules below ~30 kDa freely diffuse through the NPC, while larger molecules cannot pass the permeability barrier of the NPC, established by hydrophobic phenylalanine-glycine (FG) repeats of Nups (FG-Nups).5 For passage of larger molecules through the NPC, they must bind to nuclear transport carriers, such as importin β family members, p10/NTF2, TAP/NFX or Hikeshi that interact with FG-Nups and possess ability to translocate through NPCs (for reviews see refs.6−8).
The small GTPase Ran plays key roles in determining the directionality of nuclear transport, mediated by importin β family members, by regulating cargo binding and release.6,7 The GDP form of Ran is converted into the GTP form within the nucleus by chromatin bound Ran GTP-exchange factor (GEF) RCC1. The GTP form of Ran is converted into the GDP form by the Ran GTPase-activating protein (GAP) RanGAP1 in the cytoplasm, thereby creating a steep RanGTP concentration gradient at the boundary of the NE. For the nuclear import, importins bind to their cargoes in the cytoplasm where RanGTP concentration is low, but dissociate from the bound cargoes within the nucleus when RanGTP bind to importins. The nuclear export occurs in the opposite manner: exportins bind cargoes in the presence of RanGTP and forms a trimeric complex, but release them when RanGTP is converted into RanGDP.
In multicellular organisms, the nuclear lamina, a meshwork of type V intermediate filaments, A-type and B-type lamins, lies beneath the INM and maintains the nuclear shape and structure.9,10 Moreover, the nuclear lamina stays in contact with the cytoskeleton through its interaction with the highly conserved linker of nucleoskeleton and cytoskeleton (LINC) complex (see refs.11−13). The LINC complex consists of KASH (Klarsicht, Anc-1 and Syne homology) and Sad1 and UNC-84 (SUN) proteins, which are membrane spanning proteins that localize at the ONM and INM, respectively. Sun proteins form dimers through their coiled-coiled domain and bind KASH proteins in the perinuclear lumen via the SUN domain at their C-terminus.
At the ONM, KASH proteins interact either with actin filaments,13 microtubules,14 intermediate filaments15 or centrosomes16 via their cytoplasmic tail.
Besides the structural role, the nuclear lamina functions in gene stability and chromatin organization, which is associated with various human diseases, collectively called laminopathies.17-19
The fate of the nuclear envelope in mitosis
In the course of “open” mitosis, the NE disassembles, also referred to as nuclear envelope breakdown, and follows the fate described below.
In prophase, the permeability of the NE initially increases as NPCs disassemble in a stepwise manner upon phosphorylation by mitotic kinases such as CDK1 and Neks.20 The nuclear membrane is then partially ruptured in a process assisted by microtubule-motor proteins, dynein and its regulator dynactin21 and finally becomes completely retracted into the mitotic ER.22
Upon disassembly of the NE, the nuclear barrier dissolves and nuclear transport is annihilated. However, RanGTP gradient continues to exist around mitotic chromosomes through the action of chromatin-bound RCC1. By this way, nuclear localization signal (NLS)-containing cargoes can be recruited and released at the vicinity of mitotic chromosomes, where the concentration of RanGTP is high.23-26 Moreover, mitotic chromosomes establish a spatial environment for phosphorylation gradients, e.g., by scaffolding Aurora B kinase,27 which is the main activity of the chromosomal passenger complex (CPC) and is important for spindle assembly checkpoint and cytokinesis.28
During metaphase, most NE components are either dissolved or membrane-bound until reformation of the NE in anaphase. It was recently shown that lamin B and Nups also have important, mitotic functions.29,30
In late anaphase, NE reformation takes place on mitotic chromosomes, which serve as assembly platform for the highly controlled de novo assembly of NPCs in space and time and the reconstruction of the nuclear lamina as well as of the nuclear membrane, originating from the mitotic ER. Further, the compaction and structure of chromatin (condensed/decondensed) regulates binding of NE proteins and supports reformation of a functional NE.31
Nups, lamins and some INM proteins localize on sister chromosomes in a mutually related manner, indicating that NPC assembly and reconstruction of other NE structures involving INM proteins and lamins are coordinated. However, the molecular basis for such coordination is still very poorly understood.
Recruitment of Nuclear Membrane Proteins and Lamins at the End of Mitosis
The role of lamins in NE reformation
In mammals, one class of lamins, the A-type lamins, is derived from LMNA gene locus, which expresses the two splicing variants lamin A and lamin C. The other class of lamins is called B-type lamins, including three different proteins. Lamin B1 is encoded by the gene locus of LMNB1, and lamin B2 and B3 (B3 is only expressed in testis) are encoded by LMNB2 (reviewed by refs. 9, 10, 32 and 33). At least one form of B-type lamin is present in each cell, while expression of A-type lamins is tissue specific and regulated during development. A- and B-type lamins share the same structural elements such as a α-helical rod domain and several IgG-folds but are processed differently. Mature B-type lamins are carboxymethylated and farnesylated at the C-terminus, which favors attachment of B-type lamins to lipid membranes. Precursors of A-type lamins are modified in the same way but the last 15 amino acids are cleaved by the metalloprotease Zmpste24/FACE1, which renders mature A-type lamins membrane-unattached.10 Various human diseases like Emery-Dreifuss muscular dystrophy are related to A-type lamins and its interaction partners, but only a few diseases are known, which derive from B-type lamins.18,19
Early experiments tested the function of lamins during NE reformation in vitro with Xenopus laevis and Drosophila embryo extracts. X. laevis has three different B-type lamins (B1–3),34 whereby lamin B3 is the most abundant one. Drosophila melanogaster expresses only one B-type lamin (lamin Dm0). Immunodepletion of B-type lamins from Drosophila embryo extracts inhibited vesicle attachment to chromatin35 and chromatin decondensation in nuclear assembly assays in Xenopus egg extracts.36 Further immunodepletion of Xenopus lamin B3 gave rise to fragile and small nuclei yet with a sealed nuclear membrane,37 which were devoid of a lamina and lost their ability for DNA replication.38 In an alternative approach, Lopez-Soler and colleagues used a C-terminal peptide of Xenopus lamin B3 that bound and inhibited the function of endogenous lamin B3 and disturbed nuclear membrane targeting to chromatin, NPC formation and lamin polymerization in vitro.39 Apparent contradictions of lamin B depletion in Xenopus or Drosophila extracts could be a result of cross-reactivity of the lamin antibodies, unspecific interactions of the lamin peptide or the inefficient removal of lamin B3 from extracts. In total, these results demonstrated a role of B-type lamin in establishment of a functional nucleus. In mammalian cells, recruitment of B-type lamin to mitotic chromosomes is critical for cell survival.40,41 However, it was reported, that B-type lamin is essential for organogenesis but not for differentiation of embryonic stem cells.42 Accumulating evidences indicate that lamins have a significant, physiological role during development in various organisms,43-45 as well as in senescence,46 which might be due to their function in the maintenance of the nuclear morphology and the interaction network with other proteins at the NE that involves signaling pathways.
During NE reformation, recruitment of lamin B to mitotic chromosomes requires dephosphorylation by PP1. Steen et al.41 identified the PP1-substrate specifier AKAP (A-kinase anchoring protein) 149 as an integral membrane protein of the NE and ER membrane. In HeLa cells, when association of AKAP 149 with PP1 was inhibited, localization of B-type lamin was abolished at the end of mitosis, while it had no effect on NE localization of A-type lamin or INM proteins such as emerin and LBR. The different effect of AKAP149 on localization of B-type lamin and A-type lamin suggests that their way of assembly at end of mitosis is not interdependent and is regulated differently. Live imaging and immunofluorescent staining demonstrated that reformation of the nuclear lamina in mammalian cells occurs later than recruitment of other INM proteins like emerin, Lap2α, LBR or transmembrane Nups such as Pom121,37,47-49,59 demonstrating chromosome recruitment of some INM proteins does not depend on the assembled lamina.
INM proteins in reformation of a functional nuclear membrane
Many INM proteins can bind either or both of A- or B-type lamins.50 In addition, recruitment of INM proteins is involved in the attachment of the ER to mitotic chromosomes.22,51-53 INM proteins, which have been studied in relation to their function in NE formation, are described below.
LBR
One of the best-studied INM proteins is the lamin B receptor (p58/LBR), which was initially identified from avian erythrocytes as lamin-binding protein.54 The N-terminus of human LBR extends into the nucleoplasm, whereas the C-terminal region of LBR contains eight predicted transmembrane segments with homology to the sterol reductase ERG24 (human TM7SF2/DHCR1).55 The N-terminus interacts with chromatin,56,57 chromatin-binding proteins like heterochromatin binding protein (HP1), HA95, histone H3/4, the nuclear transport receptor importin β57 and the nucleoporins Pom12158 and ELYS/Mel28.59
There are two rare human diseases related with LBR: hydrops-ectopic calcification-“moth-eaten” (HEM)/Greenberg skeletal dysplasia and Pelger- Huët anomaly (PHA). The PHA syndrome, a hematological condition, induces hypolobulation of nuclei in granulocytes and detachment of heterochromatin from the NE.60 HEM leads to abnormalities in skeletal growth and displays parallels to a deficiency of 3β-hydroxysterol Δ(C-14) reductase, which implies that LBR has a role in cholesterol metabolism.61 Overexpression of Xenopus LBR in HeLa cells caused membrane overproduction and stack formation, suggesting a role of LBR in membrane growth.62 Whether LBR functions as sterolreductase is unclear and awaits in-depth investigation.
Multiple studies support the relevance of LBR for NE formation. LBR is required in vitro for nuclear membrane assembly in sea urchin (sea urchin homolog p56),63 rat hepatocyte or turkey erythrocyte extracts,64 and in Xenopus egg extract, depending on importin β and RanGTP.62 Depletion of hLBR in HeLa cells increased apoptotic marker caspase-3 in G1-phase65 and delayed NE formation in vivo.52 It was suggested that LBR plays a redundant function with other INM proteins (see next section).
LBR is phosphorylated by several kinases such as a SR-specific kinase and cdc2.66 Tseng et al.67 demonstrated that the mitotic phosphorylation of hLBR on residue S71 and S86 by CDK1 hindered the premature attachment of the LBR-associated nuclear membrane to chromatin, from which they concluded that mitotic phosphorylation of hLBR is important for temporal control of NE assembly.
LEM domain-containing proteins
Another group of INM proteins, which interacts with lamins and is potentially involved in NE formation, are LEM (named after INM proteins Lap2, emerin, Man1) domain containing proteins. The LEM domain, formed by 40 aa of two parallel α-helices,68,69 is shared by nonrelated INM proteins, including emerin, isoforms of lamina-associated polypeptide (Lap)2 (α, β, δ, ε, γ, ζ), Man1, Lem2/3/4 and yet uncharacterized Lem5.70 The LEM domain is specifically recognized by the protein Barrier-to-Autointegration Factor (BAF). BAF, an A-type Lamin binding protein, forms a dimer and displays DNA-looping activity, necessary for chromatin organization.71-73 The role of BAF in chromatin decondensation supports proper NE assembly.74-76 The chromatin-binding affinity of BAF is decreased after phosphorylation by VRK-1 kinase upon mitotic entry. Recently, it was reported that C. elegans protein LEM4-L and the human ortholog Lem4 inhibited the phosphorylation activity of VRK-1 in vitro and interacted with PP2A77 indicating that Lem4 regulates the BAF kinase and the phosphatase that controls the function of BAF in chromatin binding and NE reformation.
In U2OS cells, RNAi-induced depletion of the LBR, Lap2β, MAN1, BAF and the transmembrane Nups Ndc1 and Pom121 delayed reformation of a transport competent NE. Double depletion of the LBR and Lap2β enhanced the delay of NE formation, whereas exogenously expressed LBR or BAF rescued NE formation in Lap2β-depleted cells. Further, overexpression of each INM protein caused accelerated NE formation in contrast to overexpression of the ONM protein nesprin-3a,52 indicating that INM proteins and their interacting partners such as BAF have a redundant role in nuclear membrane recruitment.
Contribution of Nucleoporins to NE Formation
NPC structure
The basic molecular architecture of the NPC is well conserved among eukaryotes.78-80 Several Nups behave as subcomplexes throughout the cell cycle. Nups are divided into four groups based on their structural roles in the NPC structure (Fig. 1): transmembrane Nups, scaffold Nups, central Nups and peripheral Nups. In vertebrates, three transmembrane Nups, Pom121, NDC1 and gp210, exist, which are considered to link the scaffold structure with the pore membrane. Scaffold Nups, consisting Nup107–160 and Nup93–205 subcomplexes, build up the three ring-shaped structure for the basic body of the NPC. The Nup107–160 subcomplex forms the nucleoplasmic and cytoplasmic rings, which sandwich the central ring (also known as spoke ring), composed of the Nup93–205 subcomplex. The central transport channel, enclosed by the spoke ring, is assembled by the central Nups (Nup62, Nup58 and Nup54).81 About one third of nucleoporins, including the central Nups, possess unstructured (or disordered) phenylalanine-glycine-repeats, reaching into the central transport channel of the NPC to form the permeability barrier.82,83 Peripheral Nups organize into cytoplasmic filaments and the nuclear basket, but their exact composition is unclear and several associated factors exist at the cytoplasmic and nucleoplasmic side.84,85 The permeability barrier and peripheral Nups are important for the function of the NPC in exchanging macromolecules.86
Figure 1. Structure of the metazoan nuclear envelope (NE). Scheme of the NE. A nuclear pore complex (NPC) is inserted into the NE at places where inner nuclear membrane (INM) and outer nuclear membrane (ONM) fuse. The body of NPC consists of scaffold Nups forming three ring-like structures; cytoplasmic ring (red, cytoplasmic side: Nup107–160 complex), central ring (beige: Nup93–205 complex) and nuclear ring (red, nucleoplasmic side: Nup107–160 complex). The permeability barrier is established by central Nups (Nup62 complex). Peripheral Nups (purple) constitute the cytoplasmic fibrils and the nuclear basket. In vertebrates, there are three integral membrane pore membrane Nups (green), which are thought to anchor the NPC scaffold to the NE. Each block indicates a subcomplex of Nups. The ONM is continuous with the endoplasmic reticulum (ER), whereas the INM contains a distinct subset of INM proteins (green) that interact with the filamentous meshwork of A-type (beige) and B-type lamins (orange) beneath the INM.
Two different models of NPC assembly in interphase and in mitosis
In metazoans, NPCs are assembled twice during cell cycle: during interphase and in postmitosis.87-90 It is proposed that interphase NPC formation has different preconditions from postmitotic NPC formation: it requires insertion of nucleoporins into sealed double nuclear membranes from both sides.91 For this, ONM and INM must fuse92 and the underlying nuclear lamina must locally restructure. Furthermore, the CDK activity is necessary for NPCs assembly during interphase, but not in post-mitosis.93 Recently, it was reported that Pom121 plays a critical role for early steps of interphase NPC assembly.58,94,95
Postmitotic NPC assembly occurs in a highly temporally and spatially ordered manner.96,97 There are two favored models. The pre-NPC model suggests that formation of a pre-pore structure takes place on chromatin prior to membrane attachment and fusion.97-99 In contrast, the NPC insertion model expects that NPCs are assembled into pre-existing double membranes during postmitosis,100 which is comparable to interphase NPC formation.
In line with the pre-NPC model, it was proposed that ELYS/Mel28 is the very first Nup, which targets mitotic chromosomes via its chromatin-binding, AT-hook domain.101 ELYS/Mel28 recruits the subcomplex Nup107–160, followed by the transmembrane nucleoporin Pom121101 that interacts with the Nup93–205 complex and Nup107–160 complex.102 Some members of the Nup107–160 complex such as Nup133 contain an ALPS motif, which senses and assists binding to membrane curvature.103
The vertebrate nucleoporin ELYS/Mel28, was identified as putative transcription factor during embryogenesis in mouse.104-106 Evidences accumulated, pointing out that the major function of ELYS/Mel28 in metazoans lies at the NPC. Depletion of ELYS/Mel28 blocks postmitotic NPC formation.107-110 ELYS/Mel28 directly interacts with the Nup107–160 complex, which forms the body of NPCs and is essential for NPC formation.99 It was recently demonstrated that ELYS/Mel28 and the Nup107–160 complex are involved in regulation of the CPC,111 composed of Survivin, Borealin, INCENP and Aurora B kinase. Depletion of Seh1, a component of the Nup107–160 complex, which is responsible for targeting the Nup107–160 complex to kinetochores, caused mislocalization of Survivin and Aurora B in mitosis and induced reduction of the active kinesin MKLP1, which is an Aurora B substrate and necessary for cytokinesis.111 Similarly, depletion of ELYS/Mel28 increased the number of cells with unresolved midbody structures.108 However, mechanisms of regulation of the CPC through ELYS/Mel28 and the Nup107–160 complex are unknown. Interestingly, depletion of the nuclear basket components Nup153 and Nup50 induced a delay in the Aurora B-mediated abscission checkpoint.112 Together, these evidences indicate a connection between postmitotic NPC formation and cytokinesis through Aurora B regulation.
NE reformation through ELYS/Mel28 and other Nups
Beside the primary role of ELYS/Mel28 in postmitotic NPC formation, a function in NE formation was reported for C. elegans embryos, where reduction of MEL-28 inhibited formation of a functional NE, which had neither NPCs nor a lamina and was only partially sealed.107,113 In comparison, in vitro assembled nuclei from Xenopus egg extracts depleted of MEL-28 were smaller and devoid of NPCs but enclosed by a thin nuclear membrane.109 Interestingly, in HeLa cells, reduction of ELYS/Mel28 disturbed the recruitment and distribution of INM proteins.59 While LBR was dispersed upon ELYS/Mel28 depletion, core binding INM proteins such as emerin, BAF and Lap2α still targeted to chromosomes but were affected in their accumulation at the core region, implying a role for ELYS/Mel28 in the overall regulation of the NE architecture during postmitotic assembly.59
Other nucleoporins, which have an essential role in NE formation, are components of the Nup93–205 complex, Nup155114,115 and Nup53116-118 as well as the transmembrane pore proteins Ndc1 and Pom121.102,119,120 In Nup53-depleted Xenopus extracts, NE formation was completely inhibited.118 A truncated Nup53, which binds Nup155, in X. laevis, can restore NE formation in Nup53-depleted extracts.117 Comparably, in C. elegans, a mutated Nup53, where the mutation affects the Nup155-interacting region, demonstrated a similar defect in NE formation.116 These results indicated that the interaction of Nup53 and Nup155 is necessary for successful NE formation in C. elegans and X. laevis.116,118 Moreover, Nup155 knockout is lethal in mouse and dysfunctional Nup155 is linked to a cardiovascular disease, on the cellular level basing on decreased nuclear permeability and impaired nuclear transport,121 pointing out the significance of Nup155 for a functional NPC. Nup155 interacts with other members of the Nup93–205 complex such as Nup53 and Nup93 and also binds to Pom121 and Ndc1,102,115 suggesting that the interaction of Nup93–205 with transmembrane pore proteins is essential for NE formation. Comparably to blocking the Nup93–205 complex, reduction of Pom121 inhibited NE formation in X. laevis egg extracts as observed by TEM analysis119 and induced aberrant nuclear morphology in HeLa cells.122 Depletion of Pom121 did not only disturb the NPC assembly but decreased the protein level of Nup155, Nup53, Nup93 and Nup107,102 implying an important role of Pom121 in regulation of other Nups. The similar phenotypes observed upon depletion of Pom121, Ndc1, Nup53 or Nup155, led to the conclusion that they have a comparable function in NE assembly in invertebrates. As these Nups are essential for postmitotic NPC formation, the regulation of both processes, NPC and NE formation, appear to be closely entwined,114,119 whereby Nups play a crucial role. Evidences from higher eukaryotes confirmed the close relation between NPC and NE assembly, however, transmembrane pore proteins, Pom121 and Ndc1, displayed redundancy with other INM proteins in NE formation.52
Formation of NE subdomains
In early interphase, NPCs and lamins are unevenly distributed in cultured mammalian cells, e.g., HeLa, U2OS and IMR90 cells,123,124 human prostate cancer cell lines125 and in vivo in D. melanogaster.126 The NE region, at which the density of NPCs is high, is termed pore-rich region, in contrast to the NE region, which is devoid of NPCs, designated as pore-free island.123 Generally in human cells, the pore-rich region contains B-type lamin, Sun1, LBR and NPCs, whereas A-type lamin and binding partners such as emerin, Sun2 and BAF are accumulated at pore-free islands.123,127 These structural differences disappear during cell-cycle progression from G1 to S, and all NE components are uniformly spread at the NE in G2. Large pore-free islands were maintained in cells in which their interphase NPC formation and cell cycle progression was suppressed by a CDK inhibitor.93 This result indicated that CDK activity and consequently de novo NPC formation are necessary for the reduction of pore-free islands. Furthermore, CDK activity was required for the uniform distribution of NPCs as well as of LBR and B-type lamin, which are part of the pore-rich region. On the other hand, the de-accumulation of A-type lamin and emerin, however, was not dependent on CDK activity.93 These evidences strongly suggest that there is a relationship between NPC assembly and other NE components, especially INM proteins and lamins at the pore-rich region, during interphase. A relation between the NPC and NE components is further supported by the observation that depletion of the LINC complex protein Sun1, which can be found at the pore-rich region, induced defects in interphase NPC assembly and led to NPC clustering.94,127
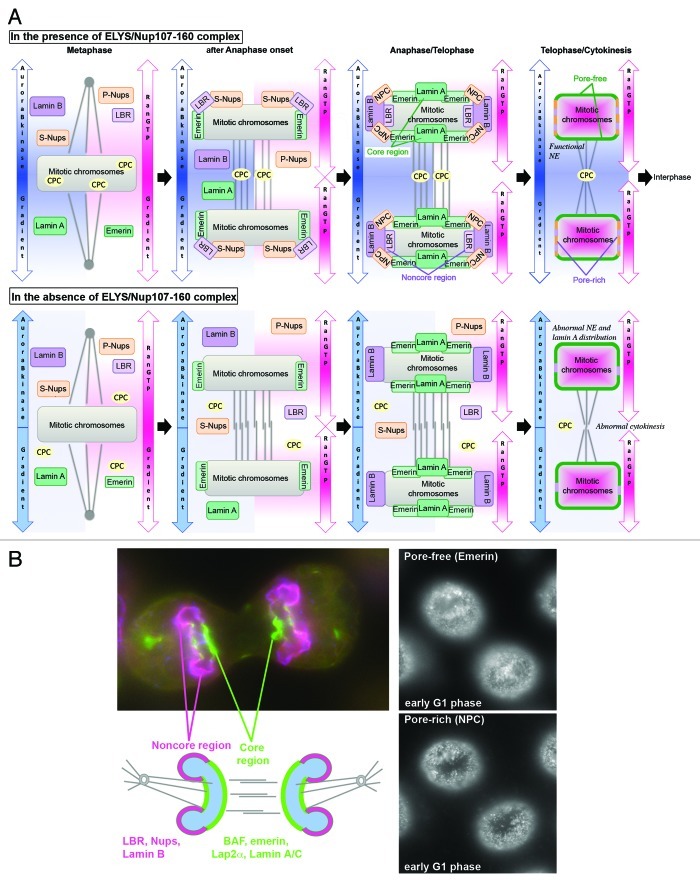
In HeLa cells, both NE subdomains, the pore-rich region and the pore-free island, are established postmitotically in telophase (Fig. 2A and B), where they correspond to the chromosomal noncore region and to the chromosomal core region, respectively.123,128 The core region is the region on chromosomes next to the spindle pole and central spindle areas, whereas the noncore regions are the peripheral regions on chromosomes surrounding the core region. The uneven distribution of NE constituents can be traced back to asymmetric targeting of A- and B-type lamins and their interacting INM proteins to telophase chromosomes. Thereby, BAF recruits LEM domain containing proteins such as emerin and Lap2α to the core region.128-130 The majority of LEM domain containing proteins specifically binds A-type lamin and A-type lamin itself requires BAF for localization at the core region.129 In contrast, B-type lamins and interacting INM proteins as well as Nups bind to the noncore regions. It was proposed that the mitotic spindle facilitates recruitment of INM proteins to the core region as microtubule (MT) disturbing chemicals diminished accumulation of proteins at the core region.129
Figure 2. Coordination of NPC and NE assembly at post-mitosis. (A) Scheme showing effects of ELYS/Nup107–160 complex on postmitotic NE/NPC assembly. In the presence of ELYS/Nup107–160 complex, scaffold Nups (Nup107–160 complex, Nup93–205 complex, also Pom121 and Ndc1), LBR and emerin, target to the noncore region of mitotic chromosomes soon after anaphase onset. During anaphase/telophase, A-type lamin binding proteins (represented by emerin) and A-type lamins accumulate at the core region of mitotic chromosomes. The core regions become the pore-free islands whereas noncore regions become the pore-rich region on the assembled, functional NE at early G1. In the absence of ELYS/Nup107–160 complex, neither scaffold Nups nor LBR can target mitotic chromosomes, although B-type and A-type lamins can localize to chromosomes in ananphase/telophase. Because absence of ELYS/Nup107–160 complex affects proper localization of the chromosomal passenger complex (CPC) that contains Aurora B kinase, the phosphorylation gradient is disrupted, which causes an aberrant distribution of core region proteins, including A-type lamins and their binding partners and also disturbs cytokinesis. RanGTP–gradient is established throughout cell cycle by chromosome bound RCC1. S-Nups, scaffold Nups; P-Nups, peripheral Nups; NPC, Nuclear Pore Complex. (B) Left: Photograph showing core region and noncore region in telophase (HeLa cell): red, LBR; green, emerin; blue, ELYS/Mel28. Right: Photographs of the nuclear surface of early G1 HeLa cells shows IF staining of emerin (upper picture), which accumulates at pore-free islands and IF staining of NPCs (lower picture), representing the pore-rich region. Pore-free islands originate from the mitotic core regions, whereas pore-rich regions derive from mitotic noncore regions as depicted in the scheme.
Conclusion
In metazoans, the overall NE architecture is established during postmitotic formation of the NE. The NE reformation requires the reassembly of the NPCs embedded into a sealed nuclear membrane, which is formed from the mitotic ER and reorganized into an ONM and INM, as well as the assembly of the nuclear meshwork of A- and B-type lamins. The coordinated sequence of reassembly during the end of mitosis poses an organizational challenge, which must be overcome to ensure the successful generation of two daughter cells with a functional nucleus. On the one hand, coordination of the NE assembly on mitotic chromosomes is dependent on the condensation/decondensation state of chromatin, which is only fragmentally understood so far. On the other hand, mitotic chromosomes itself are a center of phosphorylation and RanGTP gradients, which spatially and temporally control recruitment of NE proteins. If local chromatin changes facilitate NE subdomain formation is still an open question. Vice versa, NE subdomains could change the chromatin structure or serve as scaffold for specialized chromatin in interphase, which needs to be addressed.
Besides roles in NE reformation, Nups play important roles throughout mitosis and regulate mitotic progression. Therefore Nups are required earlier than in anaphase, although their mitotic roles must be examined in more detail.
The interplay of NE proteins during mitosis provides a basis for steady and dynamic interactions at the interphase NE. Tackling open questions about postmitotic NE reformation will give deeper insights into the control and organization of the concerted, molecular assembly of a stable NE structure in interphase, which is capable of molecular transport, signaling and chromatin regulation.
Acknowledgments
This work was supported by RIKEN Special Funding and funding awarded to N.I. by the Japan Society for the Promotion of Science (JSPS) through the Funding Program for Next Generation World-Leading Researchers (NEXT Program), initiated by the Council for Science and Technology Policy (CSTP). M. Clever thanks the RIKEN Joint Graduate School Program, International Program Associate (IPA) and the Dorothea Schlözer Fellowship for support.
Glossary
Abbreviations:
- AKAP
a-kinase anchoring protein
- BAF
barrier-to-autointegration factor
- CDK
cyclin dependent kinase
- CPC
chromosomal passenger complex
- ER
endoplasmic reticulum
- FG-repeats
phenylalanine-glycine repeats
- HP1
heterochromatin binding protein
- HEM
hydrops-ectopic calcification-“moth-eaten”/Greenberg skeletal dysplasia
- GAP
GTPase-activating protein
- GEF
GTP-exchange factor
- GTP/GDP
guanosine triphosphate/diphosphate
- INCENP
inner centromere protein
- INM
inner nuclear envelope
- KASH domain
named after proteins Klarsicht, Anc-1, Syne homology
- Lap
lamin associated polypeptide
- LBR
lamin B binding protein
- LEM domain
named after INM proteins Lap2, emerin, Man1
- LINC
linker of nucleoskeleton and cytoskeleton complex
- MKLP1
motor-kinesin like protein 1
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear envelope
- PHA
Pelger-Huët anomaly
- Pom
pore membrane protein
- PP2A
PP1, protein phosphatase 2A, protein phosphatase 1
- SR-specific kinase
serine/arginine-rich specific kinase
- SUN domain
named after proteins Sad1, UNC-84
- TM7SF2/DHCR1
transmembrane 7 superfamily member 2/dehydro-cholesterol reductase1
- VRK-1
vaccinia-related kinase-1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23796
References
- 1.Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci. 2010;123:1973–8. doi: 10.1242/jcs.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson ML. The nuclear envelope; its structure and relation to cytoplasmic membranes. J Biophys Biochem Cytol. 1955;1:257–70. doi: 10.1083/jcb.1.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–5. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 7.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–94. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imamoto N, Kose S. Heat-shock stress activates a novel nuclear import pathway mediated by Hikeshi. Nucleus. 2012;3:422–8. doi: 10.4161/nucl.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–47. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 10.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–44. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–47. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–9. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 14.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–9. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilhelmsen K, Litjens SHM, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone CJ, Misner L, Le Bot N, Tsai M-C, Campbell JM, Ahringer J, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–36. doi: 10.1016/S0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 17.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 18.Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet. 2006;7:369–405. doi: 10.1146/annurev.genom.7.080505.115732. [DOI] [PubMed] [Google Scholar]
- 19.Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–50. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/S0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 22.Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–77. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 24.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, et al. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–94. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 25.Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–5. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 26.Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, et al. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell. 2009;20:4043–58. doi: 10.1091/mbc.E09-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller BG. Self-organization of intracellular gradients during mitosis. Cell Div. 2010;5:5. doi: 10.1186/1747-1028-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 29.Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–93. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 30.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–30. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohsugi M, Adachi K, Horai R, Kakuta S, Sudo K, Kotaki H, et al. Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell. 2008;132:771–82. doi: 10.1016/j.cell.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg MW, Fiserova J, Huttenlauch I, Stick R. A new model for nuclear lamina organization. Biochem Soc Trans. 2008;36:1339–43. doi: 10.1042/BST0361339. [DOI] [PubMed] [Google Scholar]
- 33.Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2012;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- 34.Lourim D, Kempf A, Krohne G. Characterization and quantitation of three B-type lamins in Xenopus oocytes and eggs: increase of lamin LI protein synthesis during meiotic maturation. J Cell Sci. 1996;109:1775–85. doi: 10.1242/jcs.109.7.1775. [DOI] [PubMed] [Google Scholar]
- 35.Ulitzur N, Harel A, Feinstein N, Gruenbaum Y. Lamin activity is essential for nuclear envelope assembly in a Drosophila embryo cell-free extract. J Cell Biol. 1992;119:17–25. doi: 10.1083/jcb.119.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–59. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldberg MW, Jenkins H, Allen T, Whitfield WG, Hutchison CJ. Xenopus lamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J Cell Sci. 1995;108:3451–61. doi: 10.1242/jcs.108.11.3451. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steen RL, Collas P. Mistargeting of B-type lamins at the end of mitosis: implications on cell survival and regulation of lamins A/C expression. J Cell Biol. 2001;153:621–6. doi: 10.1083/jcb.153.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steen RL, Beullens M, Landsverk HB, Bollen M, Collas P. AKAP149 is a novel PP1 specifier required to maintain nuclear envelope integrity in G1 phase. J Cell Sci. 2003;116:2237–46. doi: 10.1242/jcs.00432. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, et al. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–10. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz-Böhme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–16. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guillemin K, Williams T, Krasnow MA. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat Cell Biol. 2001;3:848–51. doi: 10.1038/ncb0901-848. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–47. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–93. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhary N, Courvalin JC. Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–68. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson KL, Foisner R. Lamin-binding Proteins. Cold Spring Harb Perspect Biol. 2010;2:a000554. doi: 10.1101/cshperspect.a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–91. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–6. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- 54.Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci U S A. 1988;85:8531–4. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuler E, Lin F, Worman HJ. Characterization of the human gene encoding LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem. 1994;269:11312–7. [PubMed] [Google Scholar]
- 56.Duband-Goulet I, Courvalin J-C. Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry. 2000;39:6483–8. doi: 10.1021/bi992908b. [DOI] [PubMed] [Google Scholar]
- 57.Olins AL, Rhodes G, Welch DBM, Zwerger M, Olins DE. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus. 2010;1:53–70. doi: 10.4161/nucl.1.1.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–69. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clever M, Funakoshi T, Mimura Y, Takagi M, Imamoto N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus. 2012;3:187–99. doi: 10.4161/nucl.19595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huët anomaly) Nat Genet. 2002;31:410–4. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 61.Waterham HR, Koster J, Mooyer P, Noort Gv Gv, Kelley RI, Wilcox WR, et al. Autosomal recessive HEM/Greenberg skeletal dysplasia is caused by 3 beta-hydroxysterol delta 14-reductase deficiency due to mutations in the lamin B receptor gene. Am J Hum Genet. 2003;72:1013–7. doi: 10.1086/373938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, et al. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci. 2007;120:520–30. doi: 10.1242/jcs.03355. [DOI] [PubMed] [Google Scholar]
- 63.Collas P, Courvalin J-C, Poccia D. Targeting of membranes to sea urchin sperm chromatin is mediated by a lamin B receptor-like integral membrane protein. J Cell Biol. 1996;135:1715–25. doi: 10.1083/jcb.135.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–19. [PMC free article] [PubMed] [Google Scholar]
- 65.Lu X, Shi Y, Lu Q, Ma Y, Luo J, Wang Q, et al. Requirement for lamin B receptor and its regulation by importin beta and phosphorylation in nuclear envelope assembly during mitotic exit. J Biol Chem. 2010;285:33281–93. doi: 10.1074/jbc.M110.102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takano M, Koyama Y, Ito H, Hoshino S, Onogi H, Hagiwara M, et al. Regulation of binding of lamin B receptor to chromatin by SR protein kinase and cdc2 kinase in Xenopus egg extracts. J Biol Chem. 2004;279:13265–71. doi: 10.1074/jbc.M308854200. [DOI] [PubMed] [Google Scholar]
- 67.Tseng L-C, Chen R-H. Temporal control of nuclear envelope assembly by phosphorylation of lamin B receptor. Mol Biol Cell. 2011;22:3306–17. doi: 10.1091/mbc.E11-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin F, Blake DL, Callebaut I, Skerjanc IS, Holmer L, McBurney MW, et al. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J Biol Chem. 2000;275:4840–7. doi: 10.1074/jbc.275.7.4840. [DOI] [PubMed] [Google Scholar]
- 69.Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 2001;20:4399–407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- 71.Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278:6969–75. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- 72.Skoko D, Li M, Huang Y, Mizuuchi M, Cai M, Bradley CM, et al. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc Natl Acad Sci U S A. 2009;106:16610–5. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci U S A. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Segura-Totten M, Kowalski AK, Craigie R, Wilson KL. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J Cell Biol. 2002;158:475–85. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, et al. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J Cell Sci. 2003;116:3811–23. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- 76.Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci U S A. 2005;102:3290–5. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, et al. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–35. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 78.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–5. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 79.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 80.Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–86. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 81.Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, et al. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frey S, Richter RP, Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–7. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 83.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 84.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 85.Rothballer A, Kutay U. SnapShot: the nuclear envelope II. Cell. 2012;150:1084–1084, e1. doi: 10.1016/j.cell.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 86.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maul GG, Price JW, Lieberman MW. Formation and distribution of nuclear pore complexes in interphase. J Cell Biol. 1971;51:405–18. doi: 10.1083/jcb.51.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, et al. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci. 1999;112:2253–64. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- 89.Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–41. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maeshima K, Iino H, Hihara S, Imamoto N. Nuclear size, nuclear pore number and cell cycle. Nucleus. 2011;2:113–8. doi: 10.4161/nucl.2.2.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–3. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 92.Doucet CM, Hetzer MW. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;119:469–77. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- 93.Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol. 2010;17:1065–71. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- 94.Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584:3292–8. doi: 10.1016/j.febslet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 96.Kiseleva EV, Rutherford S, Cotter LM, Allen TD, Goldberg MW. Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci. 2001;114:3607–18. doi: 10.1242/jcs.114.20.3607. [DOI] [PubMed] [Google Scholar]
- 97.Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–65. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer MW, Galy V, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/S0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 100.Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–40. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–96. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–21. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Drin G, Casella J-F, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–46. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 104.Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, et al. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells. 2002;7:435–46. doi: 10.1046/j.1365-2443.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 105.Okita K, Nobuhisa I, Takizawa M, Ueno M, Kimura N, Taga T. Genomic organization and characterization of the mouse ELYS gene. Biochem Biophys Res Commun. 2003;305:327–32. doi: 10.1016/S0006-291X(03)00772-1. [DOI] [PubMed] [Google Scholar]
- 106.Okita K, Kiyonari H, Nobuhisa I, Kimura N, Aizawa S, Taga T. Targeted disruption of the mouse ELYS gene results in embryonic death at peri-implantation development. Genes Cells. 2004;9:1083–91. doi: 10.1111/j.1365-2443.2004.00791.x. [DOI] [PubMed] [Google Scholar]
- 107.Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–56. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 108.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–6. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, et al. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–72. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–62. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell. 2009;20:5260–75. doi: 10.1091/mbc.E09-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J Cell Biol. 2010;191:923–31. doi: 10.1083/jcb.201007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fernandez AG, Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol. 2006;16:1757–63. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- 114.Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, et al. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–31. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Busayavalasa K, Chen X, Farrants AK, Wagner N, Sabri N. The Nup155-mediated organisation of inner nuclear membrane proteins is independent of Nup155 anchoring to the metazoan nuclear pore complex. J Cell Sci. 2012;125:4214–8. doi: 10.1242/jcs.105809. [DOI] [PubMed] [Google Scholar]
- 116.Ródenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P. Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol. 2009;327:399–409. doi: 10.1016/j.ydbio.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 117.Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–94. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell. 2008;19:1753–62. doi: 10.1091/mbc.E07-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 120.Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–27. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 122.Funakoshi T, Maeshima K, Yahata K, Sugano S, Imamoto F, Imamoto N. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes. FEBS Lett. 2007;581:4910–6. doi: 10.1016/j.febslet.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 123.Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci. 2006;119:4442–51. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 124.Shimi T, Pfleghaar K, Kojima S-I, Pack C-G, Solovei I, Goldman AE, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–21. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, Shumaker DK. Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol. 2012;226:735–45. doi: 10.1002/path.3033. [DOI] [PubMed] [Google Scholar]
- 126.Furukawa K, Ishida K, Tsunoyama T-A, Toda S, Osoda S, Horigome T, et al. A-type and B-type lamins initiate layer assembly at distinct areas of the nuclear envelope in living cells. Exp Cell Res. 2009;315:1181–9. doi: 10.1016/j.yexcr.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 127.Liu Q, Panté N, Misteli T, Elsagga M, Crisp M, Hodzic D, et al. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–98. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, et al. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci. 2000;113:779–94. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- 129.Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–54. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- 130.Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, et al. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci. 2004;117:6117–28. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]