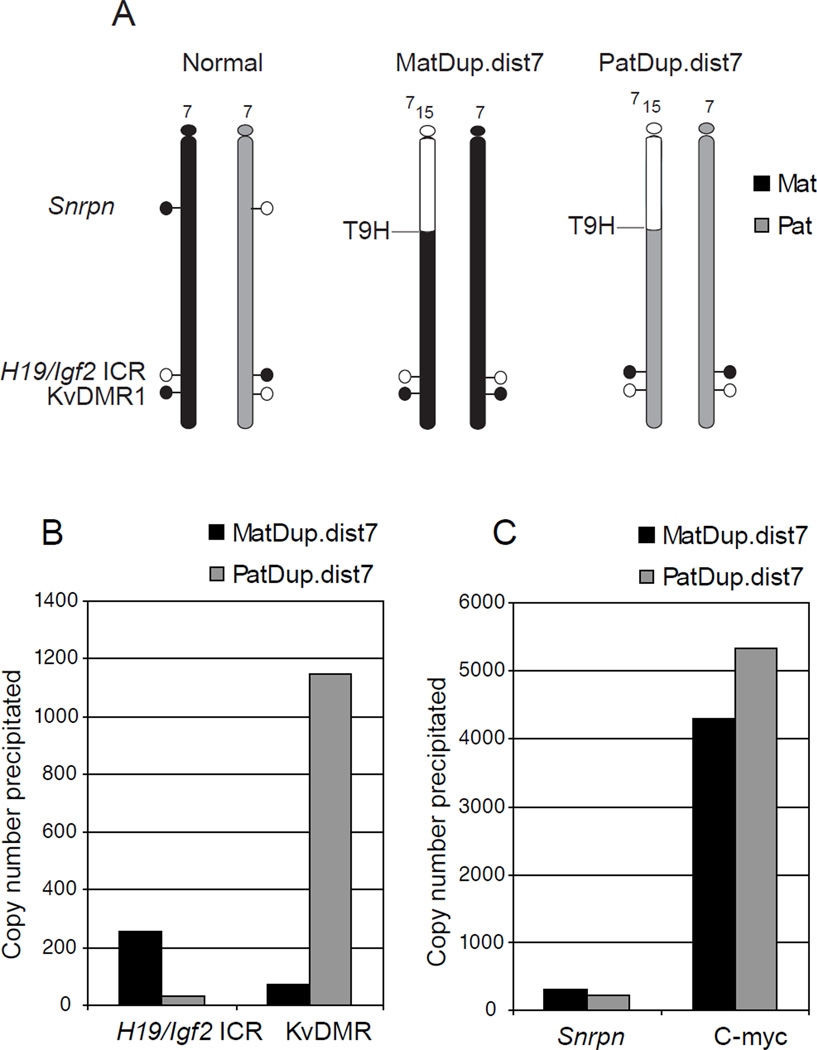
Figure 2. Detecting allele-specific H3K4me2 enrichment at imprinted regions using cells with uniparental duplications of distal Chr7.
(A) MatDup.dist7 and PatDup.dist7 MEFs carry two maternal (black) or two paternal (grey) copies of chromosome 7 regions, located distally to the T9H translocation breakpoint. These cells allow the analysis of allele-specific marks at the H19/Igf2 ICR and at the KvDMR1 along the maternally or paternally duplicated distal chromosome 7 region. Parental allele-specific methylation and hypomethylation of the DMRs is shown by closed and open lollipops, respectively. (B) Real-time PCR was used to quantify an active chromatin mark, H3K4me2, levels at specific loci in MatDup.dist7 and PatDup.dist7 MEFs at two imprinted regions. The paternally methylated H19/Igf2 ICR shows H3K4me2 in the MatDup.dist7 MEFs whereas the maternally methylated KvDMR1 exhibits H3K4me2 enrichment in PatDup.dist7 MEFs. (C) Control regions. The control Snrpn DMR is located outside of the duplicated chromosome region, therefore the allele-specificity of positive H3K4me2 enrichment can’t be discerned at this locus. The control c-myc promoter is constitutively active and enriched in H3K4me2.