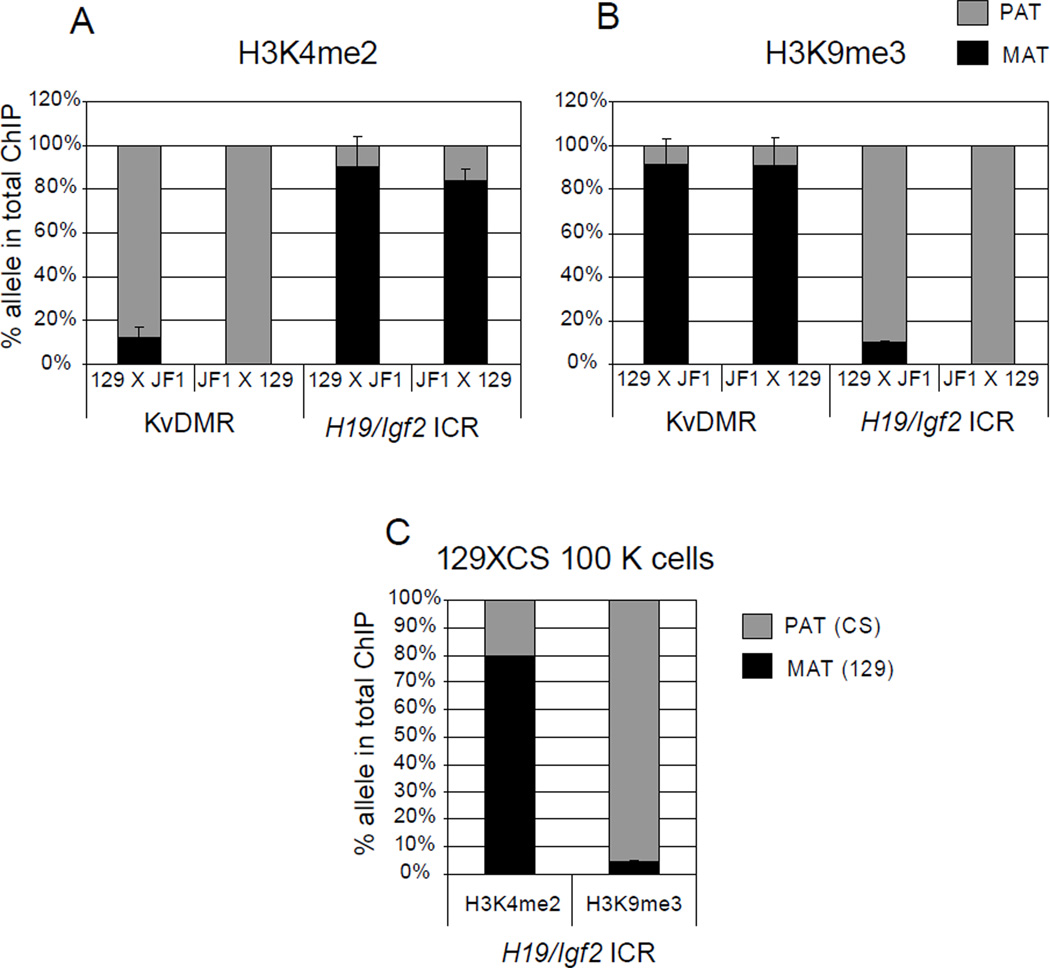
Figure 3. Detecting allele-specific chromatin in normal cells.
Chromatin was prepared from 129XJF1 (129 mother and JF1 father) and JF1X129 (JF1 mother and 129 father) MEFs and was subjected to ChIP using (A) H3K4me2 and (B) H3K9me3 antibodies. Sequenom allelotyping assays were used to measure the percent maternal and paternal component in the total ChIP DNA at the KvDMR1 and at the H19/Igf2 ICR. H3K4me2 is enriched in the unmethylated paternal allele (PAT) at the KvDMR1 and maternal allele (MAT) at the H19/Igf2 ICR. H3K9me3 shows enrichment at the reciprocal, methylated, alleles. (C) ChIP using 100000 MEFs. These MEFs were obtained by mating a 129 mother to a CAST/Ei (CS) father. Sequenom allelotyping shows correct allele specific enrichment in 129XCS MEFs for H3K4me2 and H3K9me3 at the H19/Igf2 ICR similar to ChIP from 4 µg chromatin.