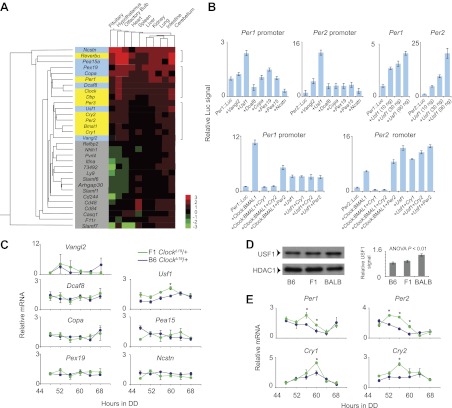
Figure 3. Usf1 is a candidate for the Soc locus.
(A) Gene expression profiling of 22 Soc candidates (gray and light blue) and nine clock genes (yellow) in 10 different tissues from Soc congenic ClockΔ19/+ mice. All tissues were collected at ZT6. The RNA copy number was calculated by a method described previously (Uno and Ueda, 2007). Copy number within all tissues for all analyzed genes was normalized (Z = mean/SD). We coded Z scores in seven different colors. Cluster analysis was performed using Systat Software. Only seven of the 22 Soc candidates (Vangl2, Usf1, Dcaf8, Copa, Pex19, Pea15 and Ncstn) show a similar expression profile with that of the clock genes within the 10 tissues examined. (B) USF1 activates Per1 and Per2 promoters. The seven Soc candidates were tested for Per1 and Per2 promoter activation (top left panels). Only USF1 activates both promoters. Each data point represents the mean ± SEM from three replicates. Activation of both Per1 and Per2 promoters by USF1 occurs in a dose-dependent manner (top right panels). USF1-mediated transcription is not inhibited by the CRY proteins. While there is strong negative inhibition by CRY of CLOCK:BMAL1-mediated transcription from the Per promoter, Per promoter activation by USF1 is not inhibited by either CRY1 or CRY2 (bottom panels). (C) Upregulation of Usf1 mRNA in liver tissue of ClockΔ19 suppressor background animals. Liver tissue was collected every 4 hr following 2 days of DD exposure. Among the seven Soc candidates tested, only Usf1 was increased in liver from F1 ClockΔ19/+ animals as determined by a two-way ANOVA (p=0.006). We did not detect an effect of time on Usf1 expression. In each panel, lines represent B6 ClockΔ19/+ (blue) and (BALB x B6)F1 ClockΔ19/+ (green) animals. Each data point represents the mean ± SEM from 2–4 mice. Asterisks indicate significant differences between ClockΔ19 suppressor and non-suppressor strain values at each time point (Tukey's post hoc, p<0.05). (D) Upregulation of USF1 protein in the liver nuclear extract from BALB/cJ genetic background. The left panel shows western blots for USF1 from B6, (BALB x B6)F1 and BALB male mouse liver samples. HDAC1 was used as a loading control. Mouse nuclear extract was prepared at ZT8. The right panel shows the normalized value for USF1 signal against HDAC1. Each bar represents the mean ± SEM from three replicates. A one-way ANOVA was significant for strain background (p=0.002). (E) Upregulation of E-box-containing circadian genes in liver from ClockΔ19 suppressor background animals. We quantified Per1, Per2, Cry1 and Cry2 mRNA levels in the same liver samples used in (C). By a two-way ANOVA, we detected significant upregulation in Per1 (p=1.5 × 10−5), Per2 (p<10−8), Cry1 (p=1.0 × 10−8) and Cry2 (p=6.6 × 10−3) transcripts. Asterisks indicate significant differences between ClockΔ19 suppressor and non-suppressor strain values at each time point (Tukey's post hoc, p<0.05).