SUMMARY
In eukaryotes, CCT is essential for the correct and efficient folding of many cytosolic proteins, most notably actin and tubulin. Structural studies of CCT have been hindered by the failure of standard crystallographic analysis to resolve its eight different subunit types at low resolutions. Here, we exhaustively assess the R-value fit of all possible CCT models to available crystallographic data of the closed and open forms with resolutions of 3.8 Å and 5.5 Å, respectively. This unbiased analysis finds the native subunit arrangements with overwhelming significance. The resulting structures provide independent crystallographic proof of the subunit arrangement of CCT, and map major asymmetrical features of the particle onto specific subunits. The actin and tubulin substrates both bind around subunit CCT6, which shows other structural anomalies. CCT is thus clearly partitioned, both functionally and evolutionary, into a substrate-binding side that is opposite to the ATP-hydrolyzing side.
INTRODUCTION
Group II chaperonins are large nanomachines that are central to protein folding in both eukaryotes and archaea. The overall structure is 16 nm in diameter and comprises two stacked rings of eight subunits each (Yébenes et al., 2011). Through a cycle that is powered by ATP hydrolysis, these subunits rotate to open and close a central folding chamber. While the archaeal systems are often homo-oligomeric (Kapatai et al., 2006), the eukaryotic chaperonin has evolved (Archibald et al., 2001) to a hetero-oligomeric complex, where each ring is made of eight paralogous subunits (CCT1 to CCT8) with a mutual sequence identity of about 30%. This divergence to eight genes occurred very early in eukaryote evolution and is highly conserved in all eukaryote species. The hetero-oligomeric nature of CCT plays a key role in CCT function by allowing for intricate substrate binding modes in the open form (Llorca et al., 2001; Spiess et al., 2006) and differential ATP hydrolysis in the closed form (Rivenzon-Segal et al., 2005; Amit et al., 2010). This functionality is tightly coupled to a precise arrangement of the eight subunits within each ring, and to a fixed relative registration of the two rings (Martín-Benito et al., 2007).
Much of the structural knowledge on group II chaperonins comes from high-resolution crystallography of archaeal systems (Ditzel et al., 1998; Shomura et al., 2004). In contrast, structural studies of CCT either by crystallography (Dekker et al., 2011; Muñoz et al., 2011) or by cryo-EM (Martín-Benito et al., 2007; Cong et al., 2010) could not achieve high enough resolution to unambiguously identify the different subunits within the electron density. Specifically, the high sequence similarity between the subunits dictates a highly similar backbone trace for all subunits. This limitation can only be overcome by resolving subunit-specific side-chains. Unfortunately, none of these studies had side-chain densities of sufficient quality to make such direct inference. As a consequence, questions pertaining to the hetero-oligomeric nature of CCT have remained unresolved, the most basic of which was that of the subunit arrangement in the native complex. This question is difficult to answer as there are eight factorial (8! = 40,320) possible arrangements of the subunits within a particle.
To resolve the subunit ambiguity, we present a new approach to low-resolution crystallography where we abandon the conventional search for specific side-chain densities with the aid of a crystallographic browser. Instead, we collectively and automatically assess the fit of many side-chains of a specific model to the crystallographic data. This automation allows us to test millions of models and to determine objectively if one of them fits the data better than others. When applied to the two crystallographic data sets of CCT that have been published to date, this approach successfully singles out with very high confidence one model for each set. These models describe how to locate the different subunits within the crystal asymmetric units and allow us to fully solve the CCT structures. The new structures not only give crystallographic proof of the native subunit arrangement of CCT, but also map various asymmetric features to specific subunits. We find that subunit CCT6-Z shows significant structural deviations from the other subunits. We also find that actin and tubulin, the main substrates of CCT, bind mainly around this subunit. The overall picture that emerges is that of a partitioned particle where specific functions such as substrate binding or ATP hydrolysis cluster to specific sides.
RESULTS
Identifying subunits in the asymmetric units
In this study we re-analyze X-ray diffraction data of the closed form of yeast-CCT at 3.8Å resolution (Dekker et al., 2011) and the open form of bovine-CCT at 5.5Å resolution (Muñoz et al., 2011). Our first aim was to assign the correct subunit type to each position in the two crystal asymmetric units. This problem is harder than determining the subunit arrangement of the particle alone, because each arrangement can fit into the electron density in eight different ways (Fig. 1A, D). There are, therefore, 2,580,480 (=8!x8x8) and 322,560 (=8!x8) possible models for the asymmetric units of the closed and open data sets, respectively. Because of the large number of possible models and the low resolution of the data, we strictly adhered to an unbiased approach that did not use calculated phases. Thus, we exhaustively built all the possible all-atom models for both asymmetric units and then measured their fit to the crystallographic data by calculating their R-values (Vaguine et al., 1999) without doing any refinement. While our fit measure is straightforward, much consideration went into the model building step. First, all our models for a particular form had exactly the same backbone without regard for the specific subunit arrangement or orientation; models differed only in the side-chain atoms added to this fixed backbone. Second, the side-chain rotamers were modeled by SCWRL4 (Krivov et al., 2009) on a backbone taken from an archaeal chaperonin (Shomura et al., 2004).
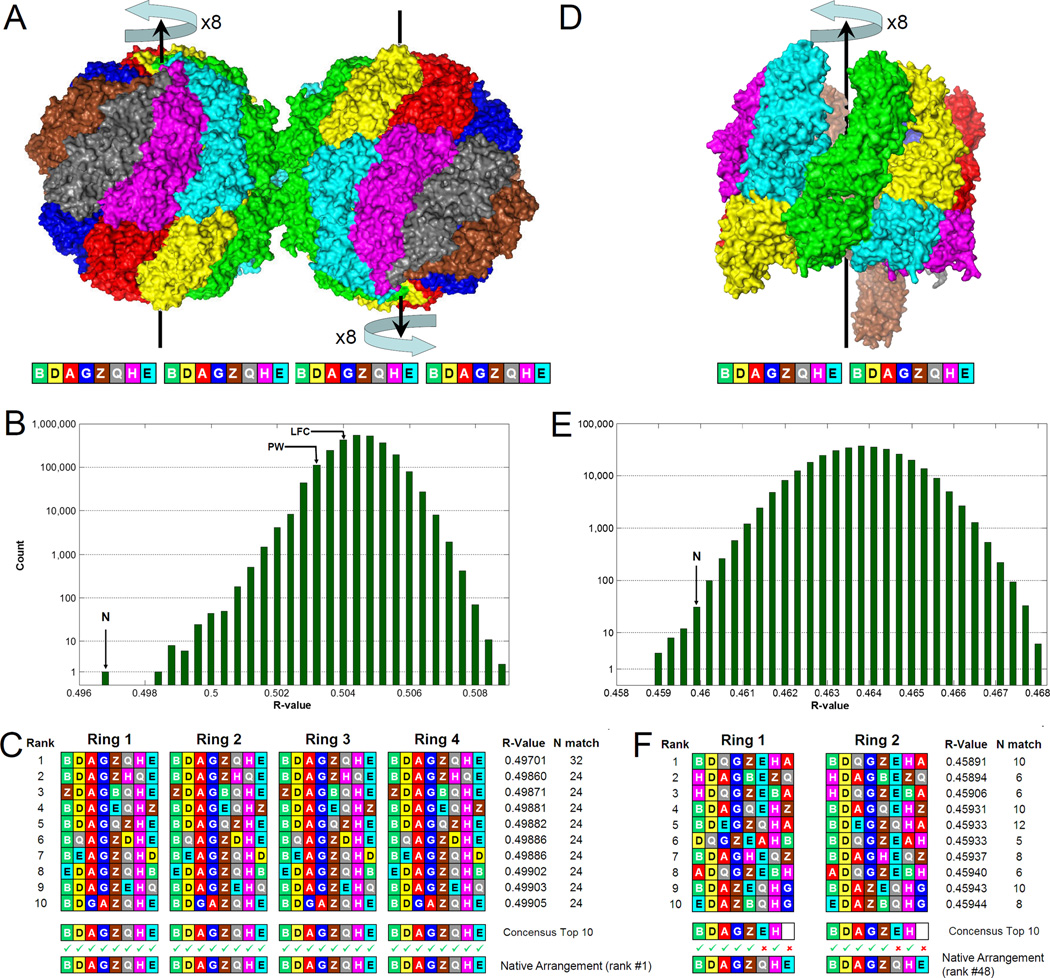
Figure 1. Identifying subunits in the asymmetric units.
(A) The asymmetric unit of the closed form contains two CCT particles, each with 2 rings of 8 subunits, colored by subunit type; they are related by a perfect non-crystallographic dyad. There are 2,580,480 (=8!×8×8) possible models of the entire asymmetric unit. Each model can be described by a 32-letter string, where each letter denotes the subunit type in a certain position. Latin letters (A,B,G,D,E,Z,H,Q) stand for subunits CCT1 to CCT8.
(B) The R-value (calculated by SFCHECK, Vaguine et al. 1999) histogram for all 2,580,480 models reveals one model (N) to best fit the crystallographic data. Previously reported subunit arrangements, PW (Dekker et al., 2011) and LFC (Cong et al., 2010), are not distinguished.
(C) A consensus string of the 10 left-most models from the histogram is identical to the model with the best R-value.
(D) The crystal asymmetric unit of the open form contains one CCT particle with 322,560 (=8!×8) possible models.
(E) The R-value histogram in this case is not as clear cut due to the lower resolution. The model (N) with the same subunit arrangement as for the closed form has a low R-value.
(F) The consensus of the 10 left-most models from the histogram is identical to the closed form consensus in 12 out of the 16 positions.
For more information about the insensitivity of the results to the method of R-value calculation, whether the structure was refined, and the subset of side chain atoms used, see Figure S1. This figure also shows that the SFCHECK R-values for the closed form have a standard deviation of just 0.0001 or 0.01%.
Without any prior information, the histogram of R-values obtained for the closed form (Fig. 1B) singles out one model of the asymmetric unit as having the best fit to the data (Rcryst=49.7%; Z-score=−10.1). This model leads by a large margin over the nearly identical second-best model (Rcryst=49.86%; Z-score=−8.0), in which the order of two subunits in the ring is swapped. In fact, the entire left tail of the distribution corresponds to models that are very similar to one another (Fig. 1C) and gives the best-R model as a consensus sequence. Independent support for this model comes from observing that its subunit arrangement is identical to that determined recently by studies that used cross-linking and mass-spectrometry (Kalisman et al., 2012; Leitner et al., 2012; Hertzog et al., 2012). For comparison, we also mark in Figure 1B the R-values of two other subunit arrangements that were previously reported. The PW model (Dekker et al., 2011) was reported for the crystallographic data used here and the LFC arrangement (Cong et al., 2010) was reported based on a cryo-EM reconstruction. These arrangements have a worse fit to the data with Z-scores of −1.6 and −0.8 for PW and LFC, respectively. Interestingly, we find that our model of the asymmetric unit reveals a perfect non-crystallographic dyad axis (perpendicular to the plane of Fig. 1A) that relates the two particles.
The dataset of the open form is much more challenging not only because of the lower resolution (5.5Å), but also because a third of the residues in one of the rings were previously reported as unstructured (Muñoz et al., 2011). Indeed, the R-value histogram (Fig. 1E) is not as clear-cut as for the closed form and there is no one model that is decisively best. We note, however, that the subunit consensus (Fig. 1F) of the top R-value models gives a particle order of BDAGZEHA-BDAGZEHA, which is identical in 12 out of the 16 subunits to the arrangement found in the closed form or by cross-linking (BDAGZQHE-BDAGZQHE). This leaves no doubt as to how to orient the previously determined subunit arrangement in the asymmetric unit. In fact, we find the corresponding model to rank among the very top models in the histogram (Rcryst=46.0%; rank #48). Observing so much side-chain signal in this very difficult crystallographic scenario is a remarkable finding. The resulting model of the asymmetric unit, assigns the full subunit in the less structured ring as CCT6-Z, and the protruding subunit in the more structured ring as CCT1-A. We note that this CCT6-Z subunit causes some steric interference in an adjacent molecule in the crystal and could be the reason for the protruding CCT1-A subunit in the latter.
Model refinement
With the subunit arrangement and orientations in the asymmetric units well confirmed, we fully refined the initial models in both forms to reveal the atomic detail needed to explain CCT function. Application of the Deformable Elastic Network (DEN) refinement (Brunger, 2007; Schröder et al., 2010) led to models with Rfree of 41.5% and 39.8% for the closed and open forms, respectively. At this point we continued to refine only the closed form structure. Further iterations of manual model building and DEN refinement led to a final structure with Rfree value of 28.37%. Complete refinement statistics for the closed and open form structures are reported in Table 1. CCT subunits contain several insertions of 5–10 residues that are subunit-specific. These insertions coincided with additional electron densities (Figs. 2A and S2) when they were structured or as clear breaks in the electron density when they were unstructured. We emphasize that these consistencies are independent support for the correctness of our subunit assignment, and were not used in the assignment process itself.
Table 1.
Crystallographic data and refinement statistics
| Crystallographic data | Closed form | Open form |
|---|---|---|
| Reference | Dekker et al., 2011 | Muñoz et al., 2011 |
| Space group | P1 | P21212 |
| Unit cell parameters | a=159.10Å | a=272.7Å |
| b=162.54Å | b=313.5Å | |
| c=268.10Å | c=158.3Å | |
| α=85.23° | ||
| β=81.15° | ||
| γ=61.17 | ||
| Wavelength (Å) | 1.00 | 1.114 |
| Resolution range (Å) | 90.0-3.8 | 100.0-5.5 |
| Completeness (%) | 91.6 (93.2)a | 92.5 (95.3)a |
| Mean I/σ(I) | 8.4 (1.92)a | 9.54 (1.2)a |
| Highest resolution shell (Å) | 4.0-3.8 | 6.0 - 5.5 |
| Refinement | ||
| Resolution range (Å) | 90.0-3.8 | 100.0-5.5 |
| No. of reflections (total / for Rfree) | 209671 / 10483 | 44462 / 2238 |
| Rwork / Rfree | 0.2479 / 0.2837 | 0.340 /0.398 |
| Average isotropic | 131.4 | 277.3 |
| B-factor (Å2) | ||
| Number of residues Protein | 32 chains | 16 chains |
| 16716 structured residues (95.5% of all residues) b | 6842 structured residues (78.8% of all residues) | |
| ADP | 32b | |
| BeF3 | 32b | |
| Mg2+ | 32b | |
| RMS deviation | ||
| Bond angle RMSD (°) | 1.62 | 0.65 |
| Bond length RMSD (Å) | 0.0128 | 0.002 |
| Ramachandran statistics (%) | ||
| Favored | 80.3 | 87.1 |
| Allowed | 15.5 | 10.7 |
| Outliers | 4.2 | 2.1 |
in the highest resolution shell.
NCS constraints enforced between the four octameric rings in the asymmetric units.
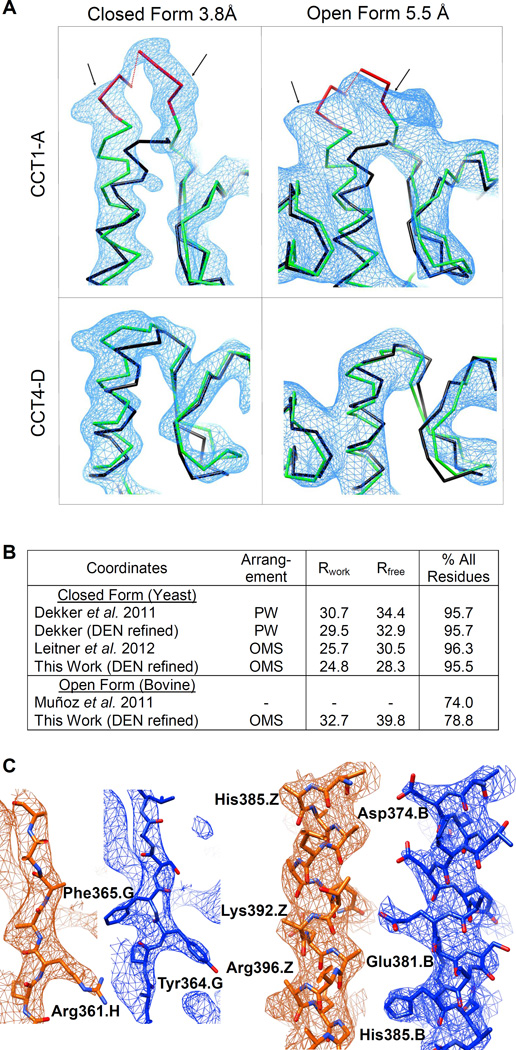
Figure 2. Refined X-ray structures.
(A) An insertion (red) unique to the sequence of the CCT1-A subunit is well resolved as an additional density (delineated by arrows) in both the closed and open datasets. Such density is not observed in the adjacent CCT4-D subunit. Overlaid for scale on the CCT traces (green) is the backbone from the archaeal 1Q3R chaperonin template (black). Density is contoured at 2σ and 1σ for the closed and open forms, respectively. See also Fig. S2.
(B) Refinement summaries for this work and for previous models of the same datasets. The PW subunit arrangement of the particle is that of Dekker et al. (2011), while the OMS arrangement (Kalisman et al., 2012) is as determined here. Over 60% of the side chains in PW are in the wrong sequence alignment position, yet the Rfree is the same as for Leitner et al. (2012).
(C) Snapshots at identical positions in the unit cell show that our model (blue) fits the m2Fo-DFc electron density map better than the model of Dekker et al. (orange). The quality of the density is also improved, showing side-chains more clearly. Maps for both models are averaged over the four rings in the asymmetric unit and contoured at 2σ. Phases are calculated from the published coordinates.
Our new closed form structure compares favorably to two other structures that were previously reported for the same dataset (Fig. 2B). The structure of Dekker et al. accompanied the initial release of the crystallographic data and was based on an incorrect subunit arrangement (PW). It is therefore not surprising that its Rfree value is 6 percentage points higher than ours and that its fit to the density is inferior (Fig. 2C). Application of DEN refinement to this structure decreased its Rfree value but not substantially. More recently, Leitner et al. reported a structure that was based on the correct subunit arrangement (OMS) and has Rfree value that is 2 percentage points higher than ours. This difference in Rfree value is likely not caused by the different core backbones, which superimpose to 0.9Å RMSD. Rather, the structures differ considerably in the trace of the chain termini and internal loops and in the rotamers of some side-chains. We note that Leitner et al. based their structure on a particle arrangement that was derived from cross-linking data, and did not systematically explore the fit of other possible arrangements or orientations to the data. We also note that their interpretation for the open form dataset is wrong, incorrectly identifying the one protruding subunit as CCT7-H instead of CCT1-A.
The weak CCT6-Z/CCT8-Q interface
The interfaces between the eight subunits in the ring are significantly different in spite of their high sequence similarity. In the closed form, the 6-Z/8-Q interface stands out as a particularly weak interface (Table 2 ; Fig. 3A), with a total buried surface area that is only 63% of the average for a ring interface (2595 Å2). Moreover, this weakness is apparent throughout the entire interface and is not limited to specific domains of the subunits. Interestingly, the second-weakest interface (8-Q/7-H) and strongest interface (3-G/6-Z) are flanking 6-Z/8-Q on both sides. We note, however, that their deviations from the average values are not nearly as drastic as that of 6- Z/8-Q, and thus might not have functional relevance.
Table 2.
Contact areas at the subunit interfaces within a ring
| Sum of Voronoi Face Area (Å2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subunit Pair | |||||||||
| Left | Right | AG | GZ | ZQ | QH | HE | EB | BD | DA |
| Equatorial | Equatorial | 844 | 899 | 460 | 770 | 572 | 717 | 761 | 710 |
| Middle | Equatorial | 642 | 642 | 369 | 504 | 358 | 571 | 608 | 392 |
| Apical | Apical | 1065 | 1252 | 796 | 911 | 1242 | 1139 | 1192 | 985 |
| Other | 303 | 283 | 47 | 220 | 620 | 271 | 189 | 472 | |
| Total | 2854 | 3076 | 1627 | 2405 | 2792 | 2698 | 2750 | 2559 | |
Surface area values are calculated from a Voronoi decomposition using the program by Gerstein and Chothia (1996). Left and right domains refer to the subunits as written in the letter pairs. The residue ranges of the domains are listed in Supplementary Table 1. Low values are shaded pink and high values are shaded green.
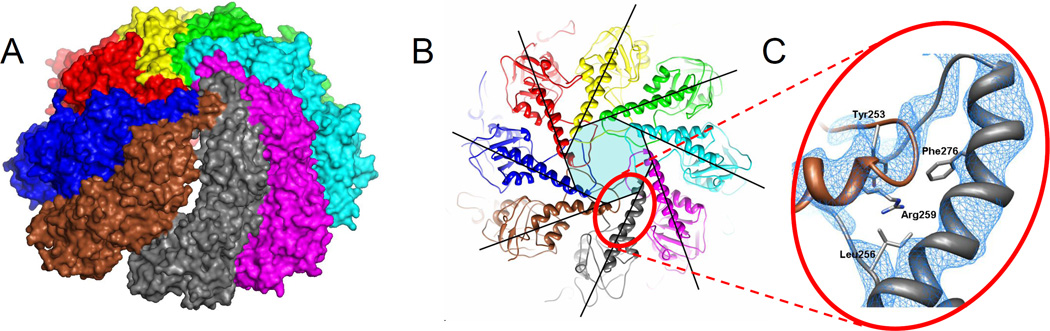
Figure 3. The weak CCT6-Z/CCT8-Q interface in the closed form.
(A) A 'crack' that occurs between these two subunits is not observed elsewhere. Only the top ring is shown. CCT6 is brown, CCT8 is grey.
(B) Top view of the closed form overlaid with a perfect 8-fold iris shows deformation of the Z capping helix. The region around Arg259 of CCT6-Z is marked.
(C) Arg259 is buried in the hydrophobic core of the cap with the guanidinium end group of the side chain exposed on the inner side of the particle. This exerts strain on the Arg259 position that is consistent with the helical deformation. In CCT6 this position is completely conserved across all eukaryotes. The corresponding positions in other subunits are always hydrophobic. The electron density map is averaged over the four rings in the asymmetric unit and contoured at 3σ. See also Fig. S3.
Between their apical domains, the 6-Z and 8-Q subunits are pushed apart because of the strain exerted by the strong kink in the helix of the 6-Z subunit that caps the folding chamber (Fig. 3B). An analysis of the evolutionary conservation at the interface (Fig. S3) implicates residue Arg259 at the tip of that helix as a probable cause of the kink. This arginine is fully conserved in the CCT6-Z subunits of all eukaryotes, but its paralogous positions in the other seven subunits are always hydrophobic and part of the hydrophobic core of the cap. The burial of the arginine sidechain in the hydrophobic core displaces the tip of the 6-Z capping helix in a direction consistent with the kink. In contrast to that example, at other positions along the 6-Z/8-Q interface the structural weakness is coupled to loss of evolutionary conservation. For example, between the equatorial domains we find Leu43 in CCT6-Z and Ser540 in CCT8-Q, which are not conserved. Yet, the paralogous positions at the other seven interfaces are highly conserved. Overall, these data strongly suggest that the weak 6-Z/8-Q interface is not a crystallographic artifact, but an intrinsic property of the closed form with functional importance. The most plausible functional explanation is that the mechanical opening of the ring starts at this interface during the transition to the open form.
The chain termini
The subunits of CCT have long N- and C-termini that are in most cases 10–20 residues long. All termini protrude from the lower part of the equatorial domains into the inner side of the particle where they were long conjectured to form a flexible 'septum' that separates the inner cavities of the two rings. We observed strong residual electron density at the midsection of the closed form that certainly supports the existence of a septum. A structural analysis of the termini in the closed form yielded similar conclusions to those reported by Dekker et al. (2011). We find that the C-termini are disordered, but the N-termini could be traced for an additional 5–10 residues outside the equatorial domain (about half their total lengths). The N-termini of all the subunits are traced inside the particle with the exception of CCT4-D whose N-terminus is instead traced to the outside of the particle through an opening between the rings. The N-termini inside the particle mainly interact with the previous subunit in the ring and in some cases with the abutting subunit on the other ring. We could not observe any common structural motif in the interactions of the N-termini with the other subunits.
In sequence, the N- and C-termini are very different. The C-termini are conserved across species both in their lengths and residue compositions, which are highly flexible and charged. The N-termini, on the other hand, are much less conserved and contain more hydrophobic residues. The lax conservation and the lack of a common structural motif lead us to believe that the structured parts of the N-termini do not carry a significant general function. Yet, a consequence of the unspecific 'stickiness' of the N-termini toward the inside surface of the particle is that they are not a significant part of the septum between the rings. It also means that it is mainly the C-termini that interact with the substrate while it is in the folding chamber. Supporting evidence for this comes from a recent cross-linking study byHerzog et al. (2012), in which the C-termini cross-linked to a substrate protein (2ABG) at about twice the rate of the N-termini. These functions of the C-termini are also consistent with their evolutionary conservation.
ATP binding and hydrolysis
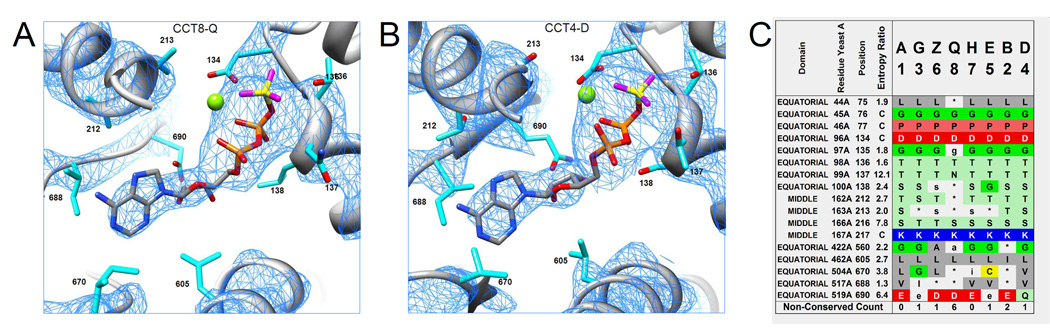
Every subunit contains a putative ATP-binding site. In the closed form, the ATP sites of all the sixteen subunits contained strong densities that fitted well to a model of ADP, BeF3 and Mg2+, which was the nucleotide analogue used to maintain the closed form. Specifically, the electron density corresponding to the adenosine and ribose moieties was well resolved in all the subunits (Fig. 4A,B). We also did not observe any significant difference in the ADP binding modes between the subunits as far as permitted by the resolution. An analysis of the evolutionary conservation of residues lining the ATP pockets (Fig. 4C) shows that they are highly conserved both across the thirteen tested species and across seven of the subunits. The exception is the CCT8-Q subunit that shows the largest variation from this consensus. For example, the GDGTT motif (positions 133 to 137) that occurs in all other subunits is mutated to GDGTN in CCT8-Q. Yet, the mutations that distinguish the ATP site of CCT8-Q from other subunits are quite conserved across species. This indicates that although the ATP site of CCT8-Q is modulated compared to other subunits, it still retains some ATP-related function. Our structure shows that part of this function is the binding (but perhaps not the hydrolysis) of ATP.
Figure 4. The ATP binding sites in the closed form.
(A and B) The ATP sites of the CCT8-Q and CCT4-D subunits show the ADP-beryllium fluoride analogues (Be-yellow; F-magenta; Mg-green). The density corresponding to the adenosine and ribose moieties is well resolved in all subunits. Side-chains are labeled according to the position indices in the conservation analysis. The density is averaged over the four rings and contoured at 3σ.
(C) The evolutionary conservation of paralogous positions lining the ATP pockets in the eight subunits. The CCT8-Q subunit shows the most variation around the ATP site compared to the other seven subunits. Positions that are identical for at least 10 of the 13 tested eukaryotic species are marked by a capital letter. Positions that are identical for at least 7 of the 13 species are marked by a lowercase letter and less conserved positions are marked by a star. High Entropy Ratio (see Methods) marks signature positions that are highly conserved across species but differ between subunit types.
Finding ATP analogues in every subunit is also surprising given previous studies (Amit et al., 2010; Jiang et al., 2011; Reissmann et al., 2012), which showed that roughly just half of the subunits are actively hydrolyzing ATP. Therefore, while the closed structure implies that all subunits are fully capable of ATP binding, we do not think it reports on ATP hydrolysis. For hydrolysis, we turn to a study by Amit et al., which used the phenotypic effects of identical mutations in the ATP sites of the different subunits as a proxy to their ATP hydrolysis potencies. When we quantify these effects (see Methods) and map them onto the ring order (Fig. 5A), we find that they cluster: the ring section comprising of subunits CCT3-G, CCT6-Z and CCT8-Q shows very little ATP hydrolysis. Very similar conclusions where also reached by a recent study (Reissmann et al., 2012) that used additional sets of mutations. In fact, the only serious discrepancy involves the classification of CCT7-H, which Reissmann et al. tagged as a weak ATP hydrolyzer. Since this subunit is between a subunit that clearly hydrolyzes ATP (CCT5-E) and a subunit that clearly hydrolyzes very little ATP (CCT8-Q), such discrepancy may arise from the different experimental setup. However, in our opinion, the strong sequence similarity of CCT7-H to the ATP-hydrolyzing subunits (see below) and the data of Amit et al. strongly suggest that CCT7-H is a potent ATP-hydrolyzer.
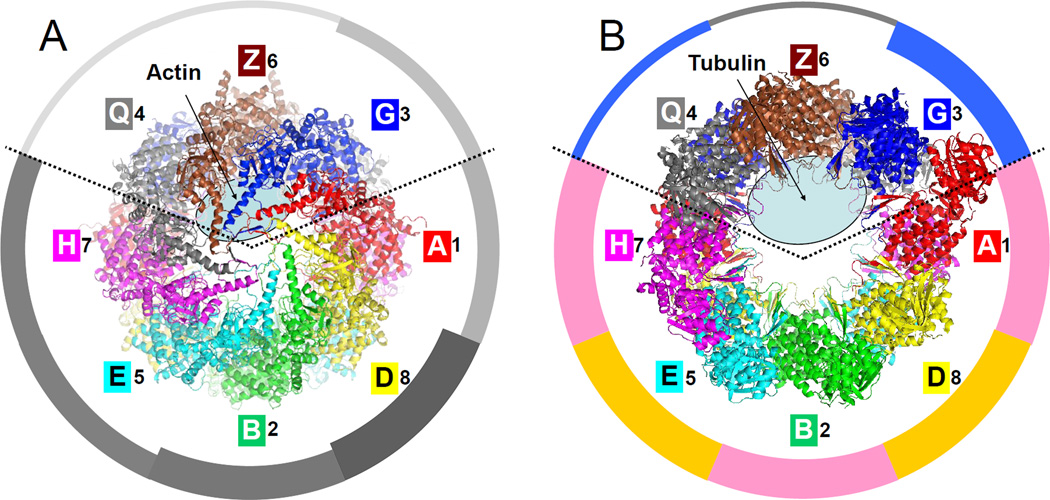
Figure 5. Substrate binding and ATP hydrolysis are partitioned in the CCT particle.
(A) Top view of the closed form with the location of residual actin density (Dekker et al., 2011) marked with an ellipse. The ATP hydrolysis potency of individual subunits (Amit et al., 2010; Methods) is proportional to width in the surrounding pie-chart.
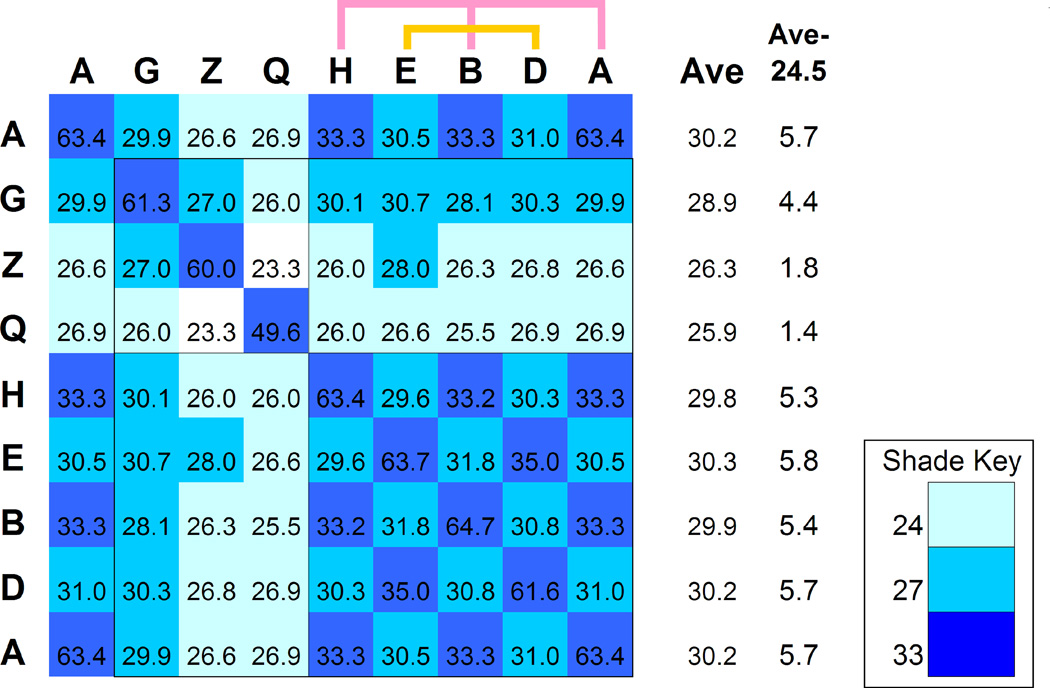
(B) Top view of the open form with the location of residual tubulin density (Muñoz et al., 2011) marked with an ellipse. The average sequence identity of each subunit to all the others is proportional to width in the pie-chart (see also Fig. 6); groups of closest sequences are colored pink (A-1, B-2, H-7) and ochre (D-4, E-5). A ring partitioning emerges: subunits 3-G, 6-Z and 8-Q are involved in substrate binding while the other subunits are involved in ATP hydrolysis.
Perhaps more problematic is the classification of CCT3-G, which according to our results is ‘halfway’ in term of ATP-hydrolysis potency. The sequence similarity to other subunits is also ambiguous in its ability to classify this subunit. Interestingly, this duality is reflected in a recent study by Nadler-Holly et al. (2012) that linked CCT3-G to the binding of Q/N-rich protein substrates, a function we attribute more to the non-hydrolyzing subunits (see below). Yet, that study also shows how this function is affected by a mutation in the ATP binding site of CCT3-G, indicating that ATP hydrolysis occurs.
Substrate binding and functional partitioning
The initial studies co-crystallized CCT with two of its main substrates: actin in the closed form (Dekker et al., 2011) and tubulin in the open form (Muñoz et al., 2011). These substrates were too disordered for any backbone tracing and were reported as residual electron densities inside the rings. Our structures now enable us to locate the specific subunits that interact with these densities (Fig. 5). In the open form, the residual tubulin density observed by Muñoz et al. is next to subunits CCT6-Z and CCT8-Q. In the closed form, we observed strong residual density inside the folding chamber next to subunits CCT3-G and CCT6-Z, which co-localizes with the density observed by Dekker et al. in their initial study. Although the closed form was co-crystallized with Plp2 in addition to actin, we follow Dekker et al. in attributing this internal density to actin alone based on three arguments. First, Plp2 was shown by cryo-electron microscopy reconstruction to bind across and above the folding chamber (Martín-Benito et al., 2004), while this density is internal. Second, photo cross-linking located Plp2 close to CCT4-D (Dekker et al., 2011), which is far from the residual density. Finally, actin is much larger than Plp2. We did not observe any other substantial density which could locate Plp2.
Overall, the picture that emerges is that of a clearly partitioned particle. Subunits CCT3-G, 6- Z and 8-Q bind the major substrates of CCT but have apparently lost their ATP-hydrolysis capabilities. They are located opposite in the ring to the five 'power-subunits', those that strongly hydrolyze ATP: CCT7-H, 5-E, 2-B, 4-D and 1-A. Of special interest is subunit CCT6-Z that appears to play a central role in substrate handling. First, at the center of the substrate binding section, it likely participates in the binding of both actin and tubulin. Second, it is involved in both the weakest and strongest intra-ring interfaces. Lastly, unlike its neighbor, CCT8-Q, its sequence is highly conserved across eukaryote species and shows sensitivity to mutations in putative substrate binding residues (Lin et al., 1997). The central role of CCT6-Z may explain why this is the only subunit for which there is an expressed isoform in mammals. It is suggestive that such isomorphism achieves the highest flexibility in substrate handling for the minimal amount of gene duplication, since the ATP-utilizing section is unaltered.
Interestingly, the functional partitioning is mirrored in the mutual sequence similarity between subunits (Figs. 5B and 6). The sequences of the five 'power-subunits' are highly similar to each other, while the sequences of the substrate-binding subunits have diverged both from them and from each other. It is beyond the scope of this work to answer whether we are observing conservation pressure due to ATP-related functions or evolutionary diversification due to substrate-related functions (or both). Nonetheless, this partitioning adds new complexity to the intriguing issue of how this hetero-oligomeric complex evolved from its homo-oligomeric archaeal ancestor.
Figure 6. Sequence identity among subunit types.
The sequence identity between the CCT subunit types was calculated using a multiple sequence alignment of 13 species. The value shown for each subunit pair is the average percent identity of all the pairwise comparisons across species. The values are shaded to emphasize the closest subunit types: with 33% identity or more in dark blue, 27% or more in cyan and 24% or more in light blue. The average off-diagonal values are given in column “Ave” and indicate how close each subunit type is to all others. The modified value in column “Ave-24.5” is used to set the width of the pie-chart segments in Fig. 5B. Subunits that are close to one another can be arranged in two sets (A, B, H) and (D, E). Subunit CCT8-Q is significantly less conserved across species.
DISCUSSION
The unbiased approach presented here confidently assigns the correct subunit type to specific electron densities in crystallographic data sets of CCT. This allows to link features in the structures to specific subunits, to match these features between structures, and to correlate other published data of CCT with the structures. The merging of these data on the two available structures of CCT shows that functions are clearly partitioned in the particle. We focus here on the implications of this partitioning on the opening of the particle. The particle opening is associated with the release of the ATP hydrolysis products (Meyer et al., 2003). If indeed the particle first opens at the CCT6-Z/CCT8-Q interface as we suggest, and since very little of these products is occurring at these subunits, then the opening signal must propagate through nearly half the ring before achieving a mechanistic effect. While such a long route is in accord with the sequential allosteric model assumed for CCT (Rivenzon-Segal et al., 2005), we cannot at present explain its benefit for CCT function. We would like to hypothesize that this mechanism somehow provides a longer lever in the conversion of the hydrolysis energy to mechanical pulling.
Our new crystallographic methodology successfully demonstrates that useful side-chain information is available from datasets at resolutions as low as 5.5 Å. This surprising achievement requires that the direct observation of side-chain densities is replaced by an unbiased and automated approach. Since we worked with unprocessed data straight off the PDB site, we strongly believe that this approach would apply for any future crystallographic study of CCT. As such, our work paves the way for more routine structural studies of CCT, which will further enhance our understanding of this elaborate system. More broadly, we foresee it as a powerful tool for general low-resolution data where conventional crystallographic approaches cannot differentiate between similar models.
EXPERIMENTAL PROCEDURES
Crystallographic datasets
This study re-analyzes the crystallographic structure factors deposited by Dekker et al. under PDB codes 3P9D and 3P9E and by Muñoz et al. under PDB code 2XSM. Their publications fully detail the complex purification and crystallization steps, which we only briefly reiterate here. For the closed form, saccharomyces cerevisiae CCT with the ANC2 mutation (G345D in CCT4; Shimon et al., 2008) was purified by using a 56-residues CBP insert after residue P374 in CCT3 (Pappenberger et al., 2006). CCT was complexed with rabbit α-actin and Plp2 cofactor, and induced into the closed conformation by incubation with ATP and beryllium fluoride. The complex was crystallized in a hanging drop in the presence of ATP and beryllium fluoride and a 3.8 Å dataset was collected (Table 1).
For the open form, CCT was purified from bovine testes and was verified by mass-spectrometry analysis to contain tubulin with the stoichiometry of one tubulin molecule per one CCT particle. CCT was incubated with ATP-γ-S, crystallized in hanging drop, and a 5.5 Å dataset was collected (Table 1).
Homology modeling of the asymmetric unit
Homology models of the individual yeast-CCT and bovine-CCT subunits were based on the template of the single repeating subunit from the archaeal thermosome of Thermococcus strain KS-1 (determined at 2.9 Å; PDB code 1Q3R), and on alignments adapted from our earlier work (Tables S2 and S3; Kalisman et al., 2012). We modeled all subunits with an identical backbone of 503 core residues by removing residues at alignment positions that had an insertion/deletion in any of the sequences. Side-chain positions were modeled by the SCWRL4 software (Krivov et al., 2009). To place a subunit model into the asymmetric unit, the model was broken into its three main domains (Table S1) and each domain was rigidly superimposed on the backbone coordinates deposited by the earlier studies (Dekker et al., 2011; Muñoz et al., 2011). The typical root mean square deviation was 0.6 Å per domain. The domain-wise model building increased the accuracy of our starting models by accounting for large variations in particle shape between thermosome and CCT. This model building protocol resulted in all-atom models for the asymmetric unit that had identical backbone coordinates regardless of their subunit composition and only differed in the coordinates of the side-chain atoms.
Refinement of the closed form model
Following the identification of the arrangement with the best crystallographic fit, we focused on its further refinement. The starting homology model had 4,024 (=8×503) residues per ring, which constituted 92.0% of its total sequence lengths (not counting the CBP insert). This model was initially refined with DEN (Brunger, 2007; Schröder et al., 2010) without imposed symmetry to Rfree = 41.48%. For the next round of refinement we introduced strong Non- Crystallographic Symmetry (NCS) restraints designed to keep the structures of the four rings similar and used torsion-angle simulated annealing. We ran 24 parallel refinements that scanned the DEN parameter space by testing all combinations of the γ-parameter (0.0, 0.2, 0.4 0.6, 0.8, 1.0) and the DEN restraint weight (1.0, 10.0, 100.0, 1000.0). The largest drop in the Rfree value to 36.9% was obtained for γ=0.2 and wDEN=1.0.
Subsequent refinement attempts were hindered by over-fitting as indicated by the tendency of Rfree to increase. We therefore switched to NCS constraints, which reduced the number of degrees of freedom four-fold and allowed further refinement to Rcryst / Rfree values of 27.06% / 30.34%, respectively. Several model building and refinement cycles were performed, with all manual modeling done in COOT (Emsley et al., 2010). A total of 4179 residues with all their constituent atoms were finally included in the ring model (95.5% of all residues). Rather than performing a costly DEN parameter search at each modeling/refinement cycle we kept the γ value at 1.0 and the weight wDEN value at 100.0.
The density in the nucleotide binding sites was not clear enough to unambiguously place the ADP molecule together with BeF3 and the Mg2+ ion. The ADP•BeF3•Mg complex was therefore restrained to maintain the coordination of BeF3 and Mg with respect to the ADP molecule as observed in the homologous site of the GroEL structure (PDB code 1SX3; determined at 2.0 Å). A restrained ENCAD (Levitt et al., 1995) run was used to improve stereo-chemistry (Fig. S4). Following the ENCAD run we replaced some of the side-chain rotamers in the subunit interface regions (see supplementary section). These optimizations resulted in a drop of 0.3% in Rfree. In the last refinement round the model quality was good enough to allow us to switch from torsion angle dynamics to Cartesian dynamics with DEN restraints. This further decreased Rcryst / Rfree to 24.79% / 28.37% to give our final structure. The effect of the DEN restraints is minimal at this stage of refinement: minimization of this final model in Cartesian coordinates without DEN restraints yields only slightly higher Rcryst / Rfree values of 25.0% / 28.5%, which indicates that the structure has been refined to a stable optimum.
Refinement of the open-form model
The starting models for the exhaustive R-value analysis included only domains that were traced in the coordinates deposited by Muñoz et al. This meant that the subunits in the top ring comprised of all three domains (equatorial, intermediate and apical), but that the subunits in the bottom ring were mostly incomplete (only one subunit had all three domains, one subunit had two domains, and the other six subunits comprised only of the equatorial domain for a total of 1142 missing residues compared to the top ring). Following the identification of the native arrangement of the asymmetric unit, we examined its electron density and found that the six missing intermediate domains in the bottom ring can all be docked into clear unassigned densities. Our starting model for refinement was therefore only missing seven apical domains in the bottom ring (total of 854 missing residues compared to the top ring). This model was refined by DEN using the default parameters. The resulting final structure had Rcryst/Rfree of 34.0% / 39.8%.
Quantification of the phenotypes in Amit et al.
That study mutated a conserved aspartic residue at the ATP pocket of each subunit in turn and observed the resulting phenotypic effects on the yeast cells (mutation D96E in CCT1-A or its equivalents in the other seven genes). Despite the relatively benign nature of this mutation the resulting phenotypic effects were drastic. Since this aspartic residue coordinates the magnesium ion at the ATP site, it is reasonable to assume that the severity of the phenotype is reporting on the extent of the ATP hydrolysis that occurs at each subunit. We quantified the severity of the phenotypes as described ahead and plotted these numbers for each subunit in Figure 5A. There were five quantifiable phenotypes measured by Amit et al.: (i) growth rate at 15°C; (ii) growth rate at 30°C; (iii) growth rate at 37°C; (iv) susceptibility to a toxic actin polymerization inhibitor and (v) susceptibility to a toxic tubulin polymerization inhibitor. For each phenotype we ranked the subunits according to the phenotype's severity going from 1 (least affected compared to WT) to 8 (most affected). Since the mutation to CCT4 was lethal, CCT4 was always given the rank of 8. The 'average phenotypic severity' of a subunit was then simply its average rank. The average rank calculated for subunits A, G, Z, Q, H, E, B, and D was 4.8, 2.8, 1.6, 1.6, 5.4, 5.4, 6.4 and 8.0, respectively.
Evolutionary conservation and sequence identity between subunits
CCT sequences for 13 species that span the eukaryote evolutionary tree were retrieved and aligned as previously described (Kalisman and Levitt, 2010). Entropy Ratio is defined as:
where ni and mki are counts of amino acid type i in all subunits or a specific subunit, k, respectively. Residues that are the same in all subunits are marked with “C” in the Entropy Ratio column.
Supplementary Material
Highlights.
CCT Subunit types can be identified in crystallographic data even at low resolutions.
Actin and tubulin both bind around CCT6.
ATP-hydrolysis and substrate binding are partitioned in the CCT particle.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM063817. Computations were done on the Bio-X2 cluster (National Science Foundation award CNS-0619926). M.L. is the Robert W. and Vivian K. Cahill Professor of Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The coordinates and structure factors of the refined structures have been deposited in the Protein Data Bank under accession codes 4AOL and 4APK for particles 1 and 2 of the closed form, respectively, and 4B2T for the open form.
SUPPLEMENTARY INFORMATION
Supplemental Information includes additional experimental procedures, three tables, and three figures and can be found with this article online.
REFERENCES
- Amit M, Weisberg SJ, Nadler-Holly M, McCormack EA, Feldmesser E, Kaganovich D, Willison KR, Horovitz A. Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J. Mol. Biol. 2010;401:532–543. doi: 10.1016/j.jmb.2010.06.037. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Blouin C, Doolittle WF. Gene duplication and the evolution of group II chaperonins: implications for structure and function. JStruct. Biol. 2001;135:157–169. doi: 10.1006/jsbi.2001.4353. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR System. Nature Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. 4.0-Å resolution cryo- EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. USA. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 2011;30:3078–3090. doi: 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel L, Löwe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowan K. Features and Development of Coot. Acta Cryst. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M, Chothia C. Packing at the protein-water interface. Proc. Natl. Acad. Sci. USA. 1996;93:10167–10172. doi: 10.1073/pnas.93.19.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmström L, Aebersold R. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Douglas NR, Conley NR, Miller EJ, Frydman J, Moerner WE. Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc. Natl. Acad. Sci. USA. 2011;108:16962–16967. doi: 10.1073/pnas.1112244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Levitt M. Insights into the intra-ring order of TRiC/CCT: A structural and evolutionary analysis. Pac. Symp. Biocomput. 2010;15:252–259. doi: 10.1142/9789814295291_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Adams CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. USA. 2012;109:2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatai G, Large A, Benesch JL, Robinson CV, Carrascosa JL, Valpuesta JM, Gowrinathan P, Lund PA. All three chaperonin genes in the archaeon Haloferax volcanii are individually dispensable. Mol. Microbiol. 2006;61:1583–1597. doi: 10.1111/j.1365-2958.2006.05324.x. [DOI] [PubMed] [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;9:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M, Hirshberg M, Sharon R, Daggett V. Potential Energy Function and Parameters for Simulations of the Molecular Dynamics of Proteins and Nucleic Acids in Solution. Computer Physics Communications. 1995;91:215–231. [Google Scholar]
- Lin P, Cardillo TS, Richard LM, Segel GB, Sherman F. Analysis of mutationally altered forms of the Cct6 subunit of the chaperonin from Saccharomyces cerevisiae. Genetics. 1997;147:1609–1633. doi: 10.1093/genetics/147.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Gómez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J. Struct. Biol. 2001;135:205–218. doi: 10.1006/jsbi.2001.4359. [DOI] [PubMed] [Google Scholar]
- Martín-Benito J, Bertrand S, Hu T, Ludtke PJ, McLaughlin JN, Willardson BM, Carrascosa JL, Valpuesta JM. Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc. Natl. Acad. Sci. USA. 2004;101:17410–17415. doi: 10.1073/pnas.0405070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Benito J, Grantham J, Boskovic J, Brackley KI, Carrascosa JL, Willison KR, Valpuesta JM. The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep. 2007;8:252–257. doi: 10.1038/sj.embor.7400894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- Muñoz IG, Yébenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Cárcer G, Malumbres M, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nature Struct. Mol. Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- Nadler-Holly M, Breker M, Gruber R, Azia A, Gymrek M, Eisenstein M, Willison KR, Schuldiner M, Horovitz A. Interactions of subunit CCT3 in the yeast chaperonin CCT/TRiC with Q/N-rich proteins revealed by high-throughput microscopy analysis. Proc. Natl. Acad. Sci. USA. 2012;13:18833–18838. doi: 10.1073/pnas.1209277109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenberger G, McCormack EA, Willison KR. Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J. Mol. Biol. 2006;360:484–496. doi: 10.1016/j.jmb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Reissmann S, Joachimiak LA, Chen B, Meyer AS, Nguyen A, Frydman J. A Gradient of ATP Affinities Generates an Asymmetric Power Stroke Driving the Chaperonin TRIC/CCT Folding Cycle. Cell Rep. 2012;2:866–877. doi: 10.1016/j.celrep.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nature Struct. Mol. Biol. 2005;12:233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A. ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J. Mol. Biol. 2008;377:469–477. doi: 10.1016/j.jmb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K. Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J. Mol. Biol. 2004;335:1265–1278. doi: 10.1016/j.jmb.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Cryst. 1999;D 55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. Chaperonins: two rings for folding. Trends Biochem. Sci. 2011;36:424–432. doi: 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.