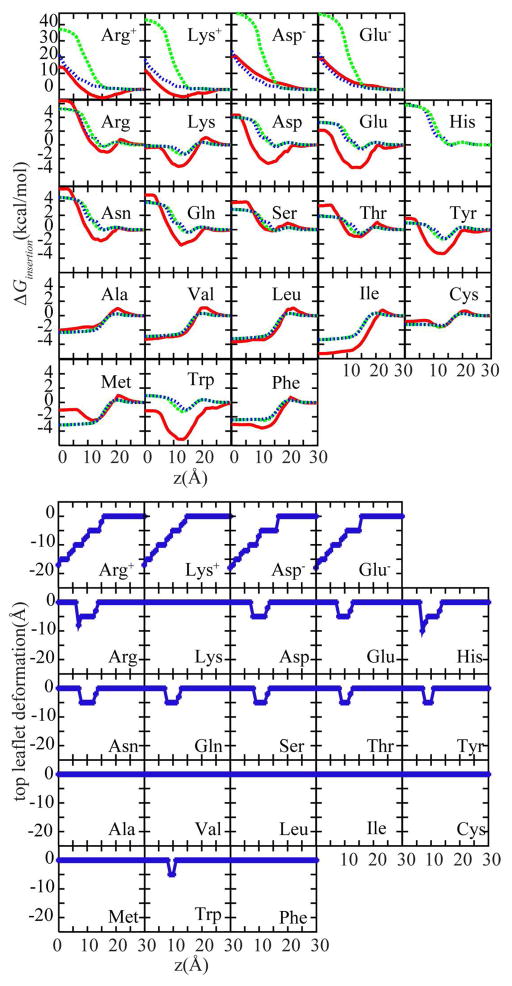
Figure 4.
Top panel: Amino acid analog insertion free energy profiles with HDGB29,34 (dashed green) and DHDGB (dotted blue) compared to results from explicit solvent/lipid simulations40 (solid red). Bottom panel: Minimized leaflet thickness upon amino acid side chain analog insertion in the DHDGB model.