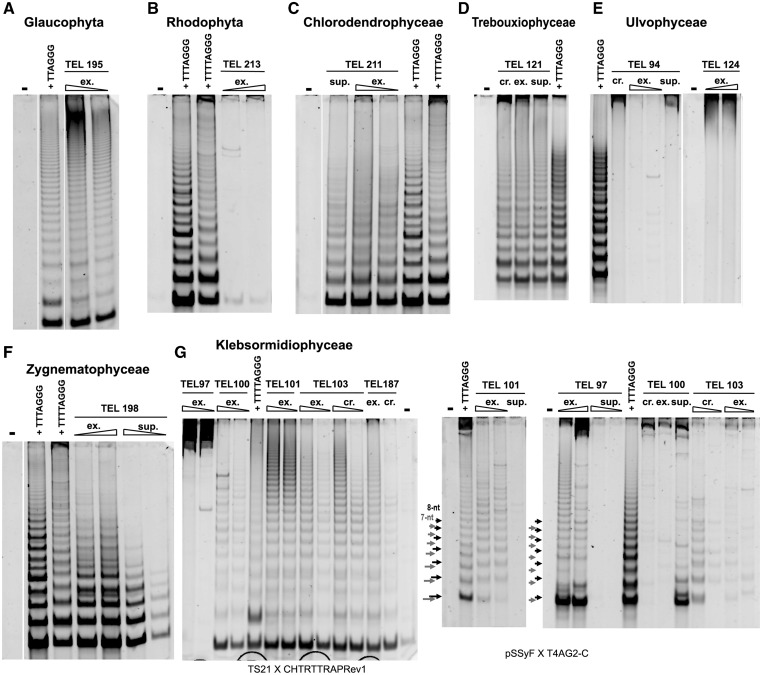
Fig. 2.—
Telomerase activity in Archaeplastida investigated by TRAP assay. Telomerase activity in representative algal strains of Glaucophyta (A, TEL195 Gla. nostochinearum), Rhodophyta (B, TEL213 R. maculata), Chlorophyta (C, TEL211 T. chui; D, TEL121 Dictyochloropsis irregularis; E, TEL94 Pseudendocloniopsis botryoides and TEL124 Pseudendoclonium basiliense), and Streptophyta (F, TEL198 Mesotaenium endlicherianum; G, TEL97 Klebsormidium subtilissimum, TEL100 K. dissectum, TEL101 K. flaccidum, TEL103 K. nitens, TEL187 K. crenulatum) grouped according to their phylogenetic provenance (indicated above panels); the activity is shown using combinations of substrate and reverse primers—GG(21) and HUTC (human-type primer) (A), 47F and TELPR30-3A (Arabidopsis-type) (B, C, F), or pSSyF and TELPR30-3A (D, E). Synthesis of telomeric repeats corresponding to the human-type and the Arabidopsis-type sequence (compare with table 1) was observed in Glaucophyta (A) and three green algal classes (C, Chlorodendrophyceae; D, Trebouxiophyceae; F, Zygnematophyceae), respectively. Negative results were obtained in Rhodophyta (B) and Ulvophyceae (E). The Klebsormidiophyceae samples (G) showed synthesis of two different telomere types; alternative combinations of the substrate primer TS21 and the Chlamydomonas-type repeat reverse primer (CHTTRAPRev1), or of the substrate primer pSSyF and the TTTTAGG-type repeat reverse primer (T4AG2-C), displayed synthesis of a 7- or an 8-nt periodicity of TRAP products (arrows) by telomerase of K. subtilissimum (TEL97) or other Klebsormidium spp., respectively (table 1). Differences in efficiency of telomerase purification during preparation from protein extracts are documented in samples shown on C, D, F, and G (summarized in supplementary table S3, Supplementary Material online). Triangles indicate different amounts of total protein (0.1 and 1 μg) in protein extract without PEG precipitation (crude, cr.), in fractions nonprecipitated (supernatant, sup) and precipitated by PEG (telomerase extract, ex), except TEL94 (E: 0.1, 0.2 μg), TEL124 (E: 0.1, 0.5 μg), TEL97 (G: 0.1, 0.8 μg), TEL100 (G: 0.1, 0.3 μg), and TEL101 (G: 0.1, 0.4 μg). When one sample is indicated, 1 μg or a higher amount of total protein mentioned earlier was used, except TEL121 (D: all 0.5 μg), TEL211 (C: sup 0.5 μg), TEL94 (E: cr. 0.1, sup 0.3 μg), and TEL101 (G: sup 0.4 μg). Telomerase-enriched extracts (50 ng of total protein) from Chlamydomonas hydra (TTTTAGGG), Arabidopsis thaliana seedlings (TTTAGGG), and Euglena stellata (TTAGGG) were used as a pattern control of an 8-, a 7-, and a 6-nt periodicity ladder, respectively; negative control (−), no extract.