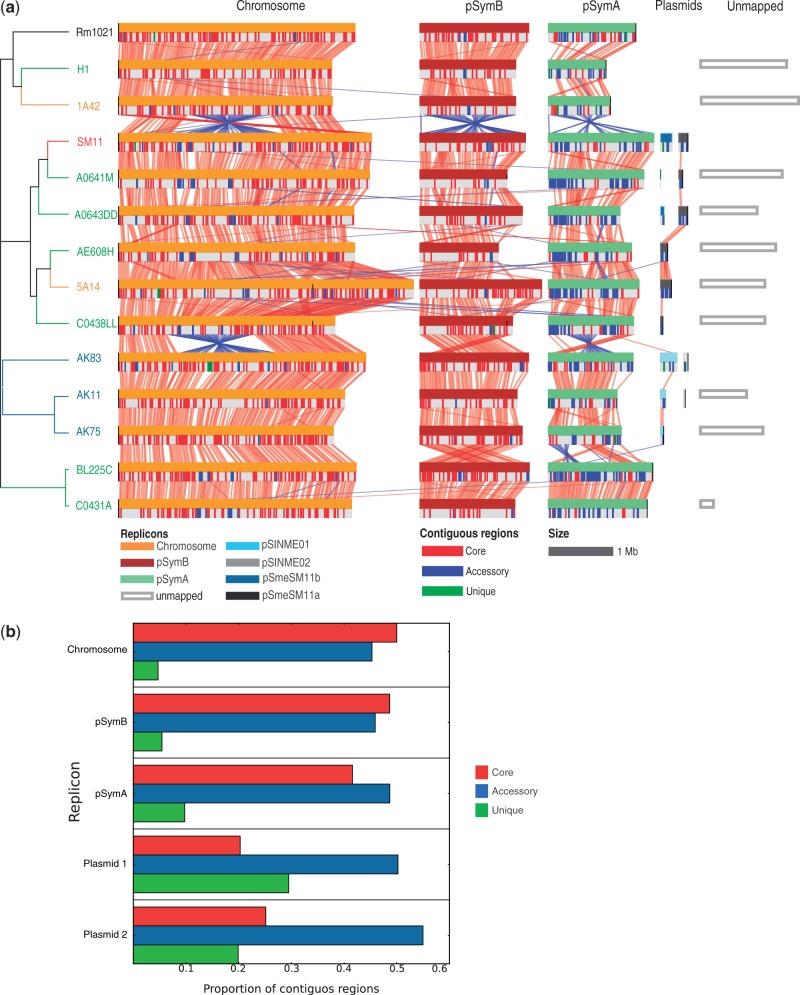
Fig. 5.—
Structural alignments of the 14 Sinorhizobium meliloti genomes. (a) Alignment between genomes is reported, following the order of the overall Bayesian dendrogram (fig. 2), the presence of core, and accessory and unique contiguous regions of orthologs whose length is over 10 kb is reported. Whole replicon inversions (as in the chromid pSymB and megaplasmid pSymA between strain 1A42 and SM11) and translocations spanning over the starting point of a replicon (as in the chromid pSymB between strain AK75 and BL225C) are artifacts dependent on the specific orientation and starting point of the nucleotide sequences. (b) Proportion of contiguous regions for each pangenomic category in each replicon.