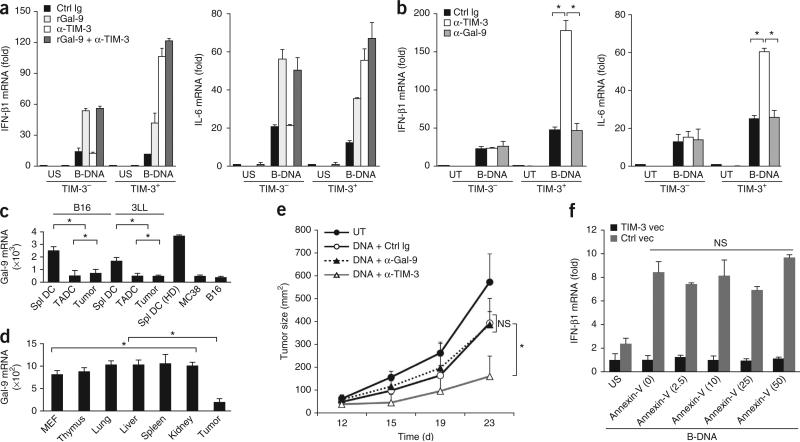
Figure 5.
TIM-3 regulates innate responses by a galectin-9-independent mechanism. (a) RT-PCR analysis of IFN-β1 and IL-6 mRNA in wild-type TIM-3+ and TIM-3– BMDCs pretreated with isotype-matched control immunoglobulin, recombinant galectin-9 (rGal-9) and/or mAb to TIM-3 (key), followed by no stimulation or stimulation for 8 h with B-DNA; results are presented relative to Actb expression. (b) RT-PCR quantification of IFN-β1 and IL-6 mRNA in TIM-3+ and TIM-3– BMDCs left untreated or treated for 24 h with B-DNA in the presence of isotype-matched control immunoglobulin, mAb to TIM-3 or mAb to galectin-9; results are presented relative to Actb expression. (c) RT-PCR quantification of galectin-9 (Gal-9) mRNA in splenic DCs (Spl DC), TADCs and tumor cells isolated from established B16 or 3LL tumors, splenic DCs from non-tumor-bearing mice (HD) and MC38 or 3LL tumor cells cultured in vitro; results are presented relative to Actb expression. (d) RT-PCR quantification of galectin-9 mRNA in MEFs, normal tissues (thymus, lung, liver, spleen and kidney) from tumor-bearing mice, and B16 tumors (Tumor); results are presented relative to expression of Gapdh (reference gene encoding glyceraldehyde phosphate dehydrogenase). (e) Tumor growth in C57BL/6 mice (n = 4 per group) left untreated or inoculated subcutaneously with B16-F10 melanoma cells (1 × 105 cells per mouse) and given intratumoral injection of control plasmid DNA in the presence of isotype-matched control immunoglobulin or mAb to galectin-9 or TIM-3. (f) RT-PCR analysis of IFN-β1 mRNA in MEFs transfected for 24 h with vector encoding TIM-3 or control vector, then left unstimulated or stimulated with B-DNA in the presence of various concentrations of recombinant annexin V (horizontal axis; in ng/ml). *P < 0.05 (paired Student's t-test). Data are representative of three experiments (a–e) or two experiments (f; error bars, s.e.m.).