Figure 7.
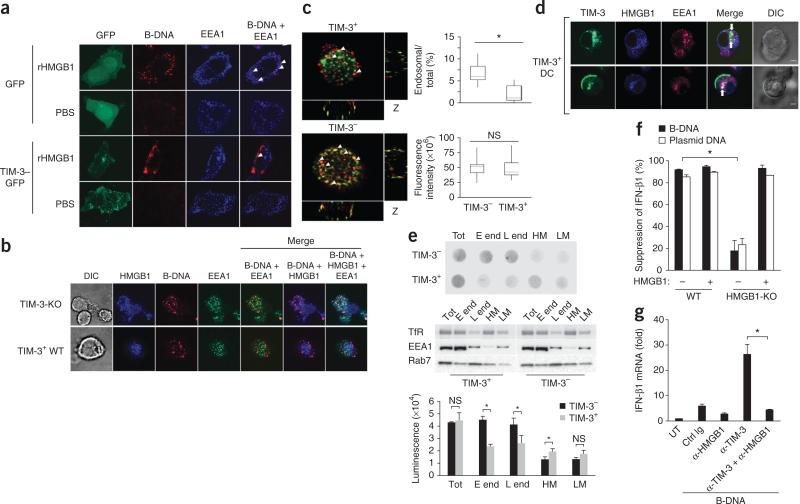
TIM-3 inhibits the recruitment of nucleic acids into endosomes. (a) Microscopy of HMGB1-deficient MEFs transfected to express green fluorescent protein alone (GFP) or green fluorescent protein–tagged TIM-3 (TIM-3–GFP), then treated with recombinant HMGB1 (rHMGB1) or PBS, assessing the localization of B-DNA (red) in EEA1+ endosomes (blue). Original magnification, ×1,000. (b) Confocal microscopy of TIM-3-deficient DCs and TIM-3+ wild-type DCs ‘loaded’ with recombinant HMGB1 (blue), then stimulated with B-DNA, assessing the localization of B-DNA (red) in EEA1+ endosomes (green). Original magnification, ×1,000. (c) Quantification of the fluorescence intensity of total cellular B-DNA (red (left); bottom right) and B-DNA in EEA1+ endosomes (green (left)) relative to total cellular B-DNA (top right) in wild-type TIM-3+ or TIM-3– BMDCs in images (left) acquired from the bottom to the top of the cells in b. Original magnification, ×1,200. (d) Confocal microscopy of the localization of TIM-3 (green) and HMGB1 (blue) in EEA1+ endosomes (red) in TIM-3+ DCs. Original magnification, ×1,000. (e) Dot-blot analysis of B-DNA (top) and immunoblot analysis of transferrin receptor (TfR), EEA1 and the late endosome marker Rab7 (middle) in total lysates (Tot), early endosomes (E end), late endosomes (L end) and heavy (HM) or light (LM) plasma membrane fractions isolated from homogenized BMDCs 2 h after treatment with B-DNA. Bottom, quantification of the dot-blot results above. (f) Suppression of IFN-β1 in wild-type or HMGB1-deficient (HMGB1-KO) MEFs transfected to express TIM-3, followed by stimulation with B-DNA or control plasmid DNA with (+) or without (–) recombinant HMGB1; results are presented relative to those of cells transfected with control vector. (g) RT-PCR analysis of IFN-β1 mRNA in TIM-3+ BMDCs left untreated or pretreated with isotype-matched control immunoglobulin or anti-HMGB1 and/or mAb to TIM-3 (horizontal axis), followed by stimulation for 8 h with B-DNA; results are presented relative to Actb expression. *P < 0.05 (paired Student's t-test). Data are representative of four experiments (a), three experiments (b,e–g) or five experiments (d) or are pooled from three separate experiments with 30 cells (c; error bars (c,e–g), s.e.m.).