Abstract
Better understanding alveolarization mechanisms could help improve prevention and treatment of diseases characterized by reduced alveolar number. Although signaling through fibroblast growth factor (FGF) receptors is essential for alveolarization, involved ligands are unidentified. FGF18, the expression of which peaks coincidentally with alveolar septation, is likely to be involved. Herein, a mouse model with inducible, lung-targeted FGF18-transgene was used to advance the onset of FGF18 expression peak, and genome-wide expression changes were determined by comparison with littermate controls. Quantitative RT-PCR was used to confirm expression changes of selected up and down regulated genes, and to determine their expression profiles in the course of lung postnatal development. This allowed identifying so far unknown target genes of the factor, among which a number are known to be involved in alveolarization. The major target was adrenomedullin, a promoter of lung angiogenesis and alveolar development, whose transcript was increased 6.9-fold. Other genes involved in angiogenesis presented marked expression increases, including Wnt2 and cullin2. Although it appeared to favor cell migration notably through enhanced expression of Snai1/2, FGF18 also induced various changes consistent with prevention of epithelial-mesenchymal transition. Together with anti-fibrotic effects driven by induction of E prostanoid receptor 2 and repression of numerous myofibroblast markers, this could prevent alveolar septation-driving mechanisms from becoming excessive and deleterious. Last, FGF18 up or down regulated genes of ECM components and epithelial-cell markers previously shown to be up or down regulated during alveolarization. These findings therefore argue for an involvement of FGF18 in the control of various developmental events during the alveolar stage.
Keywords: Animals; Animals, Newborn; Down-Regulation; drug effects; genetics; Doxycycline; pharmacology; Fibroblast Growth Factors; metabolism; Gene Expression Profiling; Gene Expression Regulation, Developmental; drug effects; Mice; Oligonucleotide Array Sequence Analysis; Pulmonary Alveoli; cytology; drug effects; growth & development; metabolism; Reverse Transcriptase Polymerase Chain Reaction; Transgenes; genetics; Up-Regulation; drug effects; genetics
Keywords: transgenic mice, alveolarization, angiogenesis, fibrosis, epithelial-mesenchymal transition.
INTRODUCTION
Lung alveolarization, the formation process of definitive alveoli, is a complex developmental event that extends from the last gestational weeks to the 2 first postnatal years. In rats and mice that are classical model animals for studying its underlying mechanisms, it is entirely postnatal; subdivision of alveolar sacs by secondary septation takes place between days 4 and 14 (although recent investigations have indicated that new alveoli can be generated later from mature septa), and septal maturation with fusion of double capillary layer into a single layer is terminated on day 20. Alveolarization involves a variety of regulatory factors, but its control is only partially understood (5). Deciphering these mechanisms is of particular importance because they are profoundly disturbed in chronic lung disease of the premature neonate also known as “new” bronchopulmonary dysplasia (BPD) (19) or in lung hypoplasia consecutive to congenital diaphragmatic hernia (CDH) (3). Furthermore, maintenance of alveolar structure necessitates the persistence of at least some of these control pathways in the adult lung, which appears to be disordered in emphysema (6).
Alveolarization is abolished in mice simultaneously devoid of fibroblast growth factor (FGF) receptors (FGFR) 3 and 4 (49). Despite normal elastic fiber gene expression in isolated lung interstitial cells, excessive elastin deposition occurs in this model with loss of typical spatial restriction, and epithelial cells contribute to the observed abnormalities (42). Thus far however, the identity of involved FGF(s) has remained elusive. The assumption that FGF18 is involved is based on the facts that (i) this mediator, similar to the most homologous other FGF-family members FGF8 and FGF17, binds FGFR4 and the IIIc-form of FGFR3 with high affinity (12, 39), and (ii) a strong rise of FGF18 expression occurs when secondary septation starts in 3 species, namely the rat (7), the mouse (8, 30) and man (3). Most strikingly, secondary septation coincides precisely with the peak of lung FGF18 expression that falls abruptly after its ending in the rat and the mouse (7, 8, 30). Indeed, FGF18 level could not be studied until completion of alveolar development in man because this occurs during the second postnatal year.
Although it is expressed at a lower rate earlier, there are evidences for FGF18 involvement in previous lung developmental stages already. Conditional expression of FGF18 in the lung of transgenic mouse fetuses from embryonic day 6 to term thus caused distal airways to adopt structural features of proximal airways with abnormal cartilage deposition, suggesting a role in the control of proximal differentiation program during pseudoglandular/canalicular stages (51). FGF18 was accordingly reported to control chondrogenesis in upper airways (11). These findings, however, do not enlighten the functional significance of the marked postnatal peak of FGF18 expression, when the tract of cartilage-containing conducting airways is complete. Another investigation evidenced reduced air space, thicker interstitial mesenchymal compartments, and presence of embedded capillaries in the prenatal lung consequently to Fgf18 gene targeting in mice, which indicates involvement at saccular stage also (45). Consistently, FGF18 expression was found to be enhanced in the stimulation of fetal rat lung growth induced by tracheal ligation (34). Unfortunately, FGF18 gene targeting cannot be used to investigate FGF18 role during alveolar stage because Fgf18−/− pups do not survive beyond birth.
Although FGF18 implication in alveolarization has therefore not yet received demonstration, it is nevertheless further supported indirectly by several observations. Thus, FGF18 stimulated ex vivo proliferation of perinatal rat lung fibroblasts and enhanced their expression of elastic fiber components (7), two events that occur during alveolar septation. Last, although FGF18 is unexplored in BPD, decreased FGF18 expression was found to be associated with impaired alveolarization in 2 instances, (i) a rat model of BPD (28) and (ii) human lung hypoplasia consecutive to CDH (3).
To document the molecular effects of FGF18 and try to explore the physiological significance of its sharp postnatal expression peak, the Tet-On system-based mouse model of conditional FGF18 expression in the lung designed by Whitsett et al. (51) was used here to provoke a rise of FGF18 in the perinatal period, hence inducing precociously an increase of expression similar to the one occurring after birth during secondary alveolar septation. Pups were collected before the onset of septation, i.e. before the rise of endogenous FGF18. This timing was chosen because we assumed that inducing the expression of the transgene later, during the stage of secondary septation, might have added little to the effects of endogenous FGF18-gene expression. In view of the considerable rise of the latter during this stage, responses are likely to be high, and even possibly maximal. Gene profiling study was then performed on genome-wide expression microarrays to identify genes up or down regulated by induced FGF18 expression. Double transgenic mouse pups bearing both the inducible FGF18 transgene and the inducer construct were compared to controls bearing only the silent FGF18 transgene. Some of the affected genes were selected on the basis of both their known functions and the extent of FGF18 effects to confirm changes by quantitative RT-PCR and to determine their expression profile during postnatal development. We thus identified as FGF18 targets a variety of genes the involvement of which in alveolarization is either demonstrated or likely, which argues for specific FGF18 functions in postnatal lung development.
MATERIAL AND METHODS
Wild type mice, transgenic mice and treatments
Wild type FVB/N mice were purchased from Charles River Laboratories (Saint Germain sur l’Arbresle, France). The transgenic mouse model has been described in detail previously (45). In brief, mice bearing the inducible construct (teto)7-CMV-FGF18 transgene were bred to SP-C-rtTA mice in which the expression of the rtTA activator construct is targeted on alveolar epithelial type II cells by the promoter of surfactant protein C. Mice bearing the transgenic constructs were bred in an FVB/N background from two male founders obtained from the Cincinnati Children’s Research Foundation. Animal experiments were performed under license from the Animal Health and Protection Service of the French Ministry of Agriculture. Because FGF18 expression in mouse lung increases postnatally to peak on days 7 to 10 (8, 30), the inducer doxycycline was given from prenatal day 2 (E18) to postnatal day 2 (P2) in order to induce earlier an expression peak similar to the one occurring during alveolar septation. Pup lungs were collected aseptically on P3 under pentobarbital anesthesia and kept frozen at −80°C until use or fixed at constant pressure for histological analysis. Experimental protocol is summarized in Fig. 1.
Fig. 1.
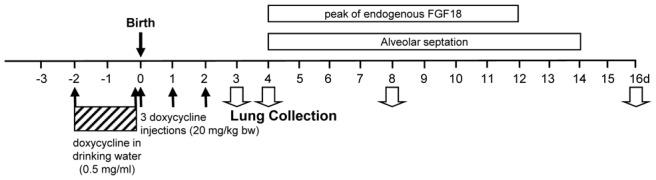
Experimental protocol for doxycycline treatment and lung sample collection. In order to induce precocious expression of FGF18, advanced as compared with endogenous peak, transgenic SP-C-rtTA- and (teto)7-FGF18-mice were bred, and their progeny (gathering both single and double transgenic individuals, and rarely wild type individuals) was treated by the inducer doxycycline. Pregnant mice received first doxycycline in drinking water (0.5mg/ml) for the 2 last prenatal days. Doxycycline was removed from drinking water on the day of birth (designated postnatal day 0 or P0), and pups received then one daily subcutaneous injection of doxycycline aqueous solution 3 mg/ml (20 mg/kg bw) on P0, P1 and P2. Their lungs were collected on P3 and kept at −80°C until RNA extraction for gene-expression analyses, or on P4, P8 or P16 for lung fixation and morphological/morphometric analyses. Post-hoc genotyping of the pups was performed on DNA extracted from tail fragment.
Lung morphology and morphometry
Methods used in this study have been described in detail previously (28). Briefly, lung fixation was performed by tracheal infusion of neutral buffered paraformaldehyde at constant 20cm H2O pressure, and fixed lung volume was measured by fluid displacement. After routine processing and paraffin embedding, 4-μm-thick mediofrontal sections through both lungs were stained with picro-indigo- carmine for morphological pictures or with hematoxylin-phloxine-saffron for morphometry. Alveolar airspace, airways, blood vessels larger than 20 μm in diameter, interstitial tissue volume densities and the alveolar surface density were determined using the point counting and mean linear intercept methods (48). Light microscope fields were quantified at an overall magnification of x440 on 20 fields per animal (10 per lung) with a systematic sampling method from a random starting point. All morphometric analyses were performed by a single observer (BCH) who was unaware of group assignment.
Genotyping and assessment of FGF18-transgene expression
Genotyping was performed on DNA extracted from mouse-tails by use of the ChargesSwitch® gDNA kit Mini Tissue kit from Invitrogen (Cergy-Pontoise, France) using the following primers for PCR in a Hybaid thermocycler: for SP-C-rtTA, 5′-GACACATATAAGACCTGGTC-3′ (forward) and (5′-AAAATCTTGCCAGRCTTCCCC-3′ (reverse); for (teto)7-FGF18, 5′-GCCATCCACGCTCTTTG-3′ (forward primer located in the CMV minimal promoter) and 5′-CAGGACTTGAATGTGCTTCCCACTG-3′ (reverse primer located in the FGF18 cDNA). The two last primers were also used in doxycycline-treated pups to assess specifically the FGF18-transgene expression response by semi-quantitative RT-PCR on total RNAs extracted from lung tissue. Quantitative RT-PCR for total FGF18 transcript was performed as described below (primers in table 1).
Table 1.
Sequence of primers used for RT-qPCR determinations
| Gene name (official symbol; accession number) | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
|
| ||
| Fibroblast growth factor 18 (FGF18; NM_008005) | TGAACACGCACTCCTTGCTAGT | GAATTCTACCTGTGTATGAACCGAAA |
| Adrenomedullin (Adm; NM_009627) | GAAGGACTTCTTTCTGCTTCAAGTG | GCGAGTGAACCCAATAACATCA |
| CD36 antigen (cd36; NM_007643) | GCAAAGTTGCCATAATTGAGTCCTA | CTGCGTCTGTGCCATTAATCA |
| Hydroxysteroid 11-beta dehydrogenase 1 (Hsd11b1; NM_007643) | TCTCTGTGTCCTTGGCCTCATA | CACTCCTCCTTGGGAGAAGCT |
| Bone sialoprotein (Ibsp1; NM_008318) | CATGCCTACTTTTATCCTCCTCTGA | AACTATCGCCGTCTCCATTTTC |
| Glutathione S transferase mu5 (Gstm5; NM_010360) | GGACGTGAAATTCAAGCTAGATCTG | CTTGTTCTTCCCGTCCATGAG |
| Ceruloplasmine (Cp; NM_007752) | TGCTGGGATGGCAACTACCT | CGTTTTCCACTTATCGCCAATT |
| Wingless-related MMTV integration site 2 (Wnt2; NM_023653) | CTGGACAGAGATCACAGCCTCTT | GCGTAAACAAAGGCCGATTC |
| Snail homolog 2 (Snai2; NM_011415) | AAGCCCAACTACAGCGAACTG | CGAGGTGAGGATCTCTGGTTTT |
| Fibulin 1 (Fbln1; NM_010180 ) | ACCAAGAAGACCCGTACCTGAA | GTCACGGCACTGCTGCTTAC |
| Prostaglandin E receptor EP2 subtype (Ptger2; NM_008964) | GCCTTTCACAATCTTTGCCTACAT | GACCGGTGGCCTAAGTATGG |
| Snail homolog 1 (Snai1; NM_011427) | GGGCGCTCTGAAGATGCA | GTTGGAGCGGTCAGCAAAAG |
| C1q and tumor necrosis factor related protein 3 (C1qtnf3; NM_030888) | GCACAACGGCAACACAGTCT | TGCATGGTTGCTGGATGTATC |
| StearoylCoA desaturase 1 (Scd1; NM_009127) | CCATGCAGGCAGGTAGTGGTA | TGCCCATGTCTCTGGTGTTTTA |
| ATP-binding cassette transporter 3 (ABCA3; AY083616) | AGCCAACATAGCAGCAGCC | TACCGAGGAGCCACGAAAAA |
| Osteoglycin (Ogn; NM_008760) | CAGATGATACATTCTGCAAGGCTAA | CCTCCAGGCGAATCTCTTCA |
| Fibroblast activation protein (Fap; NM_007986) | TGATTCTGCCTCCTCAGTTTGA | CAGACTTAACACTCTGGCTGCAA |
| Doublecortin (Dcx; NM_010025) | AAAAACTCTACACCCTTGATGGAAA | TGCATTCATTCTCATCCAAGGA |
| Cellular retinoic acid binding protein 1 (Crabp1; NM_013496) | GGGCTTCGAGGAGGAGACA | TGTGCAGTGAATCTTGTTCTCATTC |
| SRY-box containing gene (Sox2; NM_011443) | AGATGCACAACTCGGAGATCAG | GCTTCTCGGTCTCGGACAAA |
| Aldehyde dehydrogenase 1, subfamily A1 (Aldh1a1; NM_013467) | GGCTCTTCACCTGGCATCTTT | CCCATAACCAGGGACAATGTTT |
| Cytochrome P450, 1b1 (Cyp1b1; NM_009994) | AGGTTGAAGCCTTGCCAGAA | TGCTGTGGACTGTCTGCACTAA |
Generation of cRNA “target” and chip hybridization
Total RNA was extracted from lung tissue using the guanidinium isothiocynate method (TRIzol reagent, Invitrogen, Cergy-Pontoise, France), followed by purification using Rneasy columns (Qiagen, Courtaboeuf, France). Four pairs of control ((teto)7-CMV-FGF18) and double-transgenic (SP-C-rtTA, (teto)7-CMV-FGF18) pups, each pair being issued from a different litter (i.e. 4 different litters and 4 biological replicates for each condition) were selected on the basis of FGF18 transgene induction. Integrity and purity of RNA were checked by spectrophotometry and capillary electrophoresis, using the Bioanalyser 2100 and RNA 6000 LabChip kit from Agilent Technologies (Palo Alto, CA). cDNAs and biotin-labeled-cRNAs were successively synthesized using Superscript Choice system (Invitrogen) and Affymetrix IVT labeling kit (Affymetrix, Santa Clara, CA, USA), respectively. After purification, 10 μg of fragmented cRNA were hybridized to the Affymetrix GeneChip® Mouse Genome 430 2.0 Array (>39,000 transcripts). Each of the 4 control samples and of the 4 double-transgenic samples was hybridized individually on one chip (no technical replicates). Chips were automatically washed and stained with streptavidine-phycoerythrin. Arrays were scanned at 570 nm with a resolution of 1.56 μm/pixel using Affymetrix Gene Chip Scanner 3000. Expression values were generated using Microarray Suite v5.0 (MAS5, Affymetrix). Each sample and hybridization experiment underwent a quality control evaluation (table 2).
Table 2.
Evaluation of quality filter criteria in microarrays
| Sample | Scale factor | % P | Ratio 3′/5′ actin | Ratio 3′/5′ GAPDH |
|---|---|---|---|---|
|
| ||||
| (validation threshold) | Max/Min<3 | >20 and Max-Min<15 | <4 | <4 |
| D2-C | 0.504 | 60.4 | 1.25 | 0.77 |
| D4-DT | 0.582 | 59.5 | 1.30 | 0.93 |
| D34-C | 0.661 | 59.3 | 1.26 | 0.74 |
| D40-DT | 0.643 | 60.7 | 1.25 | 0.87 |
| D48-C | 0.635 | 60.6 | 1.25 | 0.78 |
| D50-DT | 0.486 | 60.8 | 1.25 | 0.81 |
| D52-C | 0.544 | 60.2 | 1.25 | 0.76 |
| D58-DT | 0.606 | 60.0 | 1.27 | 0.77 |
| Mean ± se | 0.580 ± 0.023 | 60.19 ± 0.20 | 1.26 ± 0.006 | 0.804 ± 0.004 |
Each sample and hybridization experiment underwent a quality control evaluation, including percentage of probe sets reliably detecting between 59.3% and 60.8% Present call, and 3′-5′ ratios of actin and GAPDH genes <1.3 and <1, respectively. %P: percentage of genes detected as present by Microarray Suite 5.1 software; C: control pup; DT: double-transgenic pup.
Microarray data analysis
Gene expression values were extracted from cell-probe values using the affy package (16) of the R software (15). Rma2 implementation of Robust Multiarray Averaging (17), qspline (55), pmonly and liwong (25) were used as methods of background correction, normalization, pm-correction and summarization, respectively. In order to assess replicability, the pair-wise correlations were computed between the 8 arrays. The lowest value (0.976266) indicated excellent overall replicability. Concerning intra-condition replicability, lowest values were 0.9921674 for controls and 0.979491 for double-transgenic pups. For gene selection, a one-tailed t-test for paired values was computed for both over and under-expressions. Paired t-test was used because each of the 4 double-transgenic pups was compared to its corresponding littermate control that had experienced same environment during the experiment. Because directional changes were searched for, i.e. gene expressions presenting univocal increases or decreases in all the 4 double-transgenic samples, a one–tailed t-test was used. Features were then ranked with respect to their P-value, and the list of features in the best 1% was determined. We computed the mean of the logratios, ranked the features and selected the top 0.5 for each direction of differential expression. The intersection of those 2 lists was selected for further analysis.
Reverse transcription (RT) and real-time quantitative polymerase chain reaction (qPCR)
RNAs extracted as described above were reverse-transcribed into cDNAs using 2 μg of total RNA, Superscript II reverse transcriptase, and random hexamer primers (Invitrogen) according to the supplier’s protocol. Real-time PCR was performed on ABI Prism 7000 device (Applied Biosystems, Courtaboeuf, France) using initial denaturation (10 min at 95°C), then two-step amplification program (15 s at 95°C followed by 1 min at 60°C) repeated 40 times. Melt curve analysis was used to check that a single specific amplified product was generated. Reaction mixtures consisted of 25 ng cDNA, SYBR Green 2X PCR Master Mix (Applied Biosystems), and forward and reverse primers for the various examined transcripts (displayed in table 1) in a reaction volume of 25 μl. Primers were designed using Primer Express software (Applied Biosystems). Real-time quantification was monitored by measuring the increase in fluorescence caused by the binding of SYBR Green dye to double-stranded DNA at the end of each amplification cycle. Relative expression was determined by using the ΔΔCt (threshold cycle) method of normalized samples (ΔCt) in relation to the expression of a calibrator sample used to normalize data from one plate to another, according to the manufacturer’s protocol. Each PCR run included a no-template control and a sample without reverse transcriptase. All measurements were performed in triplicates. Both 18S rRNA and the housekeeping gene Hprt1 mRNA were used as references and provided similar values (see below, Results section). For in vivo effects of FGF18-transgene induction, individual data were expressed as a percentage of the mean control value (single-transgenic pups) in each litter. For developmental expression profiles (lungs of wild type mice), data were expressed as percentage of P0 (neonates) level. Control and double-transgenic comparisons of gene-expression directional changes were made by one-tailed t test. For developmental expression profiles, considering the small sample size (4 individuals from 2 different litters per stage), non-parametric tests were used: multiple stage comparisons were made by Kruskal-Wallis analysis, followed by two-stage comparisons by Mann and Whitney U test.
RESULTS
FGF18-transgene expression and lung morphology/morphometry in doxycycline-treated mouse pups
As evaluated on P3, induction of FGF18 transgene in the lung was observed in all litters, although its rate varied widely among individuals and litters. In the 4 double-transgenic individuals selected for chip hybridization, FGF18 transgene amplification was manifest as estimated by semi-quantitative RT-PCR run with specific primers (Fig. 2). In those samples, quantification of total FGF18 expression by microarray analysis indicated 11.7-fold average increase (range: 1.5- to 20-fold), and that by RT-qPCR 6.35-fold average increase (range: 1.8- to 22-fold). In the 11 double-transgenic pups from 5 different litters used to confirm by RT-qPCR the changes evidenced by microarray data, it was similarly increased over 6-fold as compared to 10 single-transgenic littermates (2ΔΔct: 13.37 ± 2.96 vs 2.14 ± 0.16, respectively, and 641.9 ± 143.1% of controls vs 100.0 ± 6.3%, P<0.001). Although the level might have varied along treatment, this is indicative of the extent of transgene induction. Morphology of lung parenchyma appeared unaffected in double transgenic pups as compared to either wild-type or single-transgenic pups (Fig. 3). Morphometric analysis performed on pups that had undergone the same doxycycline-treatment protocol as those used for gene-expression analysis indicated no significant change in double-transgenic pups for lung volume, mean linear intercept, or alveolar surface area, either on P4 when secondary septation starts, or on P8 or P16 when it is more advanced and terminated, respectively (table 3).
Fig. 2.
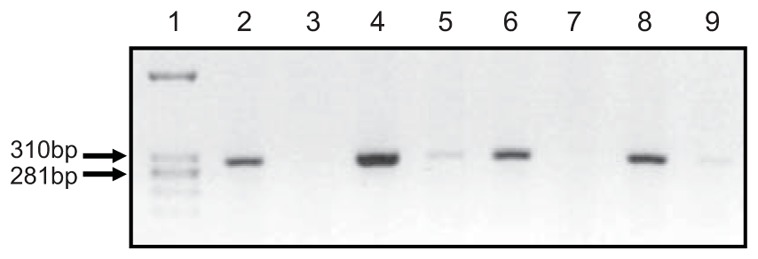
Semi-quantitative RT-PCR analysis of (teto)7-FGF18 transgene expression in the 8 mouse pup lung samples used for microarray hybridization. Transgene-specific primers were used as stated in M&M, 25 amplification cycles were run. Lane 1, molecular-size ladder; lane 2, sample D4, SP-C-rtTA/(teto)7-FGF18 (double transgenic), litter 1; lane 3, sample D2, (teto)7-FGF18 (single-transgenic control), litter 1; lane 4, sample D34, SP-C-rtTA/(teto)7-FGF18, litter 2; lane 5, sample D40, (teto)7- FGF18, litter 2; lane 6, sample D48, SP-C-rtTA/(teto)7-FGF18, litter 3; lane 7, sample D50, (teto)7- FGF18, litter 3; lane 8, sample D52, SP-C-rtTA/(teto)7-FGF18, litter 4; lane 9, sample D58, (teto)7-FGF18, litter 4. FGF18-transgene expression was manifest in double-transgenic pups bearing both the inducible FGF18 transgene and the activator construct whereas it was not or barely detectable in their littermates bearing only the FGF18 transgene.
Fig. 3.
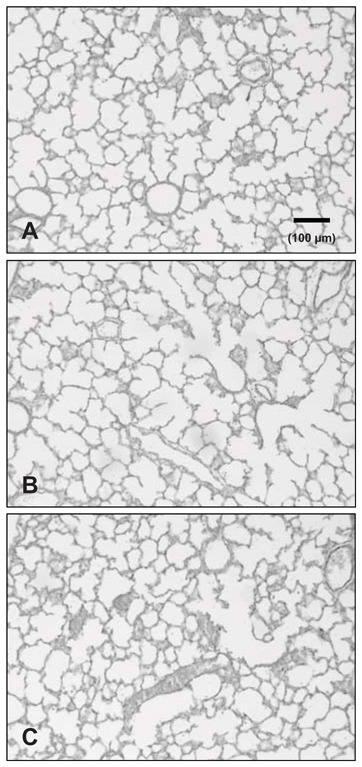
Morphological aspects of mouse lungs on postnatal day 4. Pups were issued from a same litter and were treated by doxycycline as stated in M&M and Fig. 1. Lungs were collected on P4 that revealed as the earliest stage for adequate pressure fixation in this species. Lungs were fixed with paraformaldehyde at constant pressure, embedded in paraffin, cut at 4μm and stained by picro-indigo- carmine. A. Wild-type mouse pup. B double-transgenic pup (SP-C-rtTA/(teto)7-FGF18). C. Single-transgenic pup ((teto)7-FGF18). Morphology was similar in the 3 instances.
Table 3.
Morphometric analysis of the lungs of doxycycline-treated pups
| P4 | P8 | P16 | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| n | controls 10 | double transgenic 9 | controls 8 | double transgenic 8 | controls 14 | double transgenic 8 |
| VL (cm3) | 0.135 ± 0.009 | 0.152 ± 0.010 | 0.249 ± 0.014 | 0.271 ± 0.017 | 0.376 ± 0.015 | 0.392 ± 0.027 |
| Sva (cm2/cm3) | 294 ± 12 | 277 ± 10 | 340 ± 18 | 293 ± 17 | 361 ± 12 | 358 ± 17 |
| Vvp (%) | 0.964 ± 0.007 | 0.986 ± 0.019 | 0.989 ± 0.002 | 0.992 ± 0.002 | 0.995 ± 0.001 | 0.992 ± 0.002 |
| MLI (μm) | 13.8 ± 0.5 | 14.6 ± 0.5 | 12.0 ± 0.7 | 14.0 ± 0.8 | 11.3 ± 0.4 | 11.3 ± 0.5 |
| Sa (cm2) | 38 ± 3 | 41 ± 2 | 83 ± 6 | 79 ± 7 | 135 ± 7 | 139 ± 10 |
Treatment with doxycycline was the same as that for animals used for microarray analysis. Pups were collected from 3 or 4 litters per stage at postnatal days 4, 8 or 16. Data are mean ± SE. Abbreviations: n, number of individuals per group; VL, lung volume; Sva, alveolar surface density; Vvp, volumetric density of lung alveolar parenchyma; MLI, mean linear intercept; Sa, absolute surface area of airspaces. No significant difference was observed between the 2 groups for any parameter either at P4, P8 or P16 (two-tailed t-test).
Array data analysis
Primary microarray data are available at the Gene Expression Omnibus Database (www.ncbi.nih.gov/geo/ GEO accession number: GSE22283). Statistical analysis retrieved 487 genes and EST significantly up-regulated, and 1235 genes and EST significantly down-regulated by FGF18 induction. Only characterized genes with defined function(s) were considered further. Fold-change of the 77 up-regulated genes ranged from 1.12-fold to 4-fold their respective basal expression levels in single-transgenic controls. Maximum expression decrease of down regulated genes was about 70%. In order to avoid exceedingly large tables and discussion about limited expression changes only genes with expression increased or decreased at least 25% are displayed in tables 4 and 5, respectively (i.e. 32 up-regulated genes including five genes of particular interest increased 22.4 to 24.2%, and 104 down-regulated genes).
Table 4.
Results of array analysis: genes up regulated at least 25%* by FGF18 induction in the 4 double-transgenic mouse pups as compared to their respective littermate single-transgenic controls (one-tailed paired t test).
| Accession # | Name | Funct. Cat. | log2-fold change | fold change | P value |
|---|---|---|---|---|---|
| NM_009627 | adrenomedullin (Adm) | Signal, Angio | 2.02628 | x 4.074 | 0.0494 |
| NM_007643 | CD36 antigen | Lmetab+Antifib | 1.06464 | x 2.092 | 0.0463 |
| NM_008288 | hydroxysteroid 11-beta dehydrogenase 1 (Hsd11b1) | Enz, Ster metab | 1.00447 | x 2.0 | 0.0364 |
| NM_008318 | bone sialoprotein (integrin-binding sialoprotein, IBSP) | ECM+Recept | 0.83998 | x 1.79 | 0.014 |
| NM_010360 | glutathione S-transferase, mu 5 (Gstm5) | Enz, Antiox | 0.75917 | x 1.693 | 0.0379 |
| NM_007752 | ceruloplasmin (Cp) | Enz, Antiox | 0.72622 | x 1.654 | 0.0393 |
| NM_023653 | wingless-related MMTV integration site 2 (Wnt2) | Signal, Angio+Migr | 0.58788 | x 1.5 | 0.0357 |
| NM_029796 | leucine-rich alpha-2-glycoprotein 1 (Lrg1) | Antiox | 0.55743 | x 1.472 | 0.0028 |
| NM_021282 | cytochrome P450, 2e1, ethanol inducible (Cyp2e1) | Enz, Antiox | 0.55580 | x 1.47 | 0.0305 |
| BI964347 | a disintegrin and metalloproteinase domain 12 (meltrin alpha) | Enz, ECM | 0.54291 | x 1.457 | 0.0283 |
| NM_033037 | cysteine dioxygenase 1, cytosolic (Cdo1) | Enz, Antiox | 0.51247 | x 1.426 | 0.037 |
| AF333251 | Th2-specific cytokine FISP/IL24 | Signal, Antifib | 0.48965 | x 1.4 | 0.0417 |
| NM_010023 | dodecenoyl-Coenzyme A delta isomerase (Dci) | Enz, Lmetab | 0.48113 | x 1.4 | 0.0077 |
| C77655 | CD84 antigen | Immun | 0.46537 | x 1.381 | 0.0311 |
| NM_011415 | snail homolog 2 (Drosophila) (Snai2) | Signal, Migr+EMT | 0.45841 | x 1.374 | 0.0412 |
| NM_010180 | fibulin 1 (Fbln1) | ECM+Antifib | 0.45225 | x 1.368 | 0.0354 |
| BB131147 | Borg4-pending/binder of Rho GTPase 4 | Recept, Antifib | 0.44920 | x 1.365 | 0.0261 |
| BQ176864 | protein phosphatase 1, regulatory (inhibitor) subunit 3C (Ppp1r3c) | Enz, Glyc metab | 0.43991 | x 1.357 | 0.0408 |
| BC028307 | Similar to sushi-repeat protein | Migr | 0.43074 | x 1.348 | 0.0257 |
| NM_010717 | LIM-domain containing, protein kinase (Limk1) | Enz, Antifib | 0.42335 | x 1,341 | 0,006 |
| NM_011146 | peroxisome proliferator activated receptor gamma (Pparg) | Signal, Lmetab | 0.41266 | x 1.331 | 0.0425 |
| NM_008118 | gastric intrinsic factor (Gif) | Transport | 0.38563 | x 1.306 | 0.0403 |
| NM_008059 | G0G1 switch gene 2 (G0s2) | Prolif | 0.37920 | x 1.3 | 0.0334 |
| X14607 | lipocalin 2/Mouse SV-40 induced 24p3 mRNA | Migr | 0.35332 | x 1.277 | 0.0197 |
| NM_011611 | tumor necrosis factor receptor superfamily, member 5 (Tnfrsf5) | Signal, Immun | 0.33640 | x 1.263 | 0.0425 |
| NM_023323 | brix domain containing 1 (Bxdc1) | Prolif | 0.33067 | x 1.258 | 0.0237 |
| BC008277 | Similar to Endothelin receptor type A | Recept, Angio | 0.32332 | x 1.251 | 0.0207 |
| NM_008964 | prostaglandin E receptor EP2 subtype (Ptger2) | Recept, Antifib | 0.31244 | x 1.242 | 0.0427 |
| NM_019389 | chondroitin sulfate proteoglycan 2 (Cspg2) | ECM | 0.30793 | x 1.238 | 0.0234 |
| NM_080849 | NIMA-related expressed kinase 8 (Nek8) | Enz, Prolif | 0.29273 | x 1.225 | 0.0356 |
| NM_007996 | adrenodoxin | Antiox | 0.29229 | x 1.225 | 0.0264 |
| NM_011427 | snail homolog 1, (Drosophila) (Snai1) | Signal, Migr+EMT | 0.29147 | x 1.224 | 0.0213 |
Five genes of interest increased 22.4 to 24.2% have been added to the list.
Abbreviations: Funct. Cat., functional category of gene product; Angio, angiogenesis; Antifib, anti-fibrotic effects; Antiox, anti-oxidant metabolism; ECM, extracellular-matrix component or metabolism; EMT, epithelial-mesenchymal transition; Enz, enzyme or enzyme inhibitor; Glyc metab, glycogene megtabolism; Immun, immune system; Lmetab, lipid metabolism; Migr, cell migration; Prolif, cell proliferation; Recept, receptor; Signal, signaling molecule; Ster metab, steroid metabolism; Transport, transporter molecule.
Genes with name and symbol in bold character had changes checked by RT-qPCR.
Table 5.
Results of array analysis: genes down regulated at least 25% by FGF18 induction in the 4 double-transgenic mouse pups as compared to their respective littermate single-transgenic controls (one-tailed paired t test).
| Accession # | Name | Funct. Cat. | log2-fold change | fold change | P value |
|---|---|---|---|---|---|
| NM_030888 | C1q and tumor necrosis factor related protein 3 (C1qtnf3/cartonectin) | Signal, Lipog | −1.59897 | x 0.33 | 0.0094 |
| NM_030601 | Ca(2+)-sensitive chloride channel 2 (Cacc) | Transport, Epm | −1.39070 | x 0.381 | 0.0039 |
| NM_009127 | stearoyl-Coenzyme A desaturase 1 (Scd1) | Enz, Lipog | −1.28176 | x 0.411 | 0.0088 |
| AY083616 | ATP-binding cassette transporter ABCA3 | Transport, Epm | −1.05604 | x 0.481 | 0.0004 |
| NM_008760 | osteoglycin (Ogn) | ECM | −1.04551 | x 0.484 | 0.0088 |
| NM_007986 | fibroblast activation protein (Fap/seprase) | Enz, ECM, Migr | −1,02764 | x 0.491 | 0.0002 |
| AK007703 | homolog to ATP-binding cassette transporter, sub-family A, member 3 | Transport, Epm | −0.97681 | x 0.508 | 0.0003 |
| NM_010025 | doublecortin | Migr | −0.94387 | x 0.52 | 0.0103 |
| NM_021400 | proteoglycan 4 (Prg4) | ECM | −0,92209 | x 0.528 | 0.0187 |
| NM_009181 | sialyltransferase 8 (alpha-2, 8-sialytransferase) B (Siat8b) | Enz, Glycoconj | −0.89706 | x 0.537 | 0.0265 |
| NM_009608 | actin, alpha, cardiac (Actc1) | MCm | −0.88653 | x 0.541 | 0.0397 |
| NM_009393 | troponin C, cardiac/slow skeletal (Tncc1) | MCm | −0.86216 | x 0.55 | 0.0262 |
| NM_013496 | cellular retinoic acid binding protein I (Crabp1) | Retino metab | −0.85952 | x 0.551 | 0.0373 |
| NM_011674 | UDP-glucuronosyltransferase 8 | Enz, Detox | −0.82905 | x 0.563 | 0.0247 |
| NM_013808 | cysteine-rich protein 3 (Csrp3) | MCm | −0.80843 | x 0.571 | 0,0064 |
| NM_010867 | myomesin 1 (Myom1) | MCm | −0.79768 | x 0.575 | 0.0049 |
| AF311699 | c-type lectin protein MT75 (chondrolectin) | ECM | −0.79070 | x 0.578 | 0.0003 |
| NM_015825 | SH3-binding domain glutamic acid-rich protein (Sh3bgr) | MCm | −0.78637 | x 0.581 | 0.0121 |
| NM_033268 | actinin alpha 2 (Actn2) | MCm | −0.77289 | x 0.585 | 0.0405 |
| NM_013834 | secreted frizzled-related sequence protein 1 (Sfrp1) | Prolif, EMT | −0.76298 | x 0.589 | 0.0005 |
| NM_010858 | myosin light chain, alkali, cardiac atria (Myla) | MCm | −0.75605 | x 0.592 | 0.0373 |
| NM_018870 | phosphoglycerate mutase 2 (Pgam2) | Enz, Inter metab | −0.74443 | x 0.597 | 0.0268 |
| NM_011620 | troponin T3, skeletal, fast (Tnnt3) | MCm | −0.73578 | x 0.6 | 0.0170 |
| NM_011443 | high mobility group box protein (SRY-box containing gene 2, Sox2) | Trancrip, Epm | −0.72852 | x 0.6 | 0.0028 |
| NM_013467 | aldehyde dehydrogenase family 1, subfamily A1 (Aldh1a1) | Retino metab | −0.71960 | x 0.607 | 0.0216 |
| NM_011111 | plasminogen activator inhibitor, type II (Serpinb2) | Signal, Migr | −0.71960 | x 0.607 | 0.0019 |
| NM_016689 | aquaporin-3 | Transport, Epm | −0.69277 | x 0.619 | 0.0330 |
| NM_009944 | cytochrome c oxidase subunit VIIa-H precursor (Cox7ah) | Enz, Detox | −0.68876 | x 0.62 | 0.0211 |
| NM_011395 | solute carrier family 22 (organic cation transporter), member 3 (Slc22a3) | Transp | −0.68500 | x 0.622 | 0.0149 |
| NM_010856 | Mouse alpha cardiac myosin heavy chain (Myhca) | MCm | −0.67333 | x 0.627 | 0.0422 |
| NM_009922 | calponin 1 (Cnn1) | MCm | −0.66432 | x 0.631 | 0.0093 |
| BB193413 | aquaporin 4 (Aqp4) | Transport, Epm | −0.65006 | x 0.637 | 0.0335 |
| AF274959 | uteroglobin related protein 1A/secretoglobin (Scgb3a2) | Transport, Epm | −0.64771 | x 0.638 | 0.0196 |
| NM_008973 | pleiotrophin | Sign | −0.64583 | x 0.639 | 0.0007 |
| AF128849 | cytochrome P450 2B10 related protein (Cyp2b20) | Enz, Detox | −0.63401 | x 0.644 | 0.0318 |
| NM_010356 | glutathione S-transferase, alpha 3 (Gsta3) | Enz, Detox | −0.63049 | x 0.646 | 0.0207 |
| NM_033597 | myeloblastosis oncogene (Myb) | Transcript | −0.61924 | x 0.65 | 0.0174 |
| NM_021396 | butyrophilin-like protein (Btdc) | Immun | −0.61661 | x 0.652 | 0.0013 |
| NM_008182 | glutathione S-transferase, alpha 2 (Yc2) (Gsta2) | Enz, Detox | −0.61598 | x 0.652 | 0.0069 |
| NM_009405 | troponin I, skeletal, fast 2 (Tnni2) | MCm | −0.61569 | x 0.653 | 0.0417 |
| NM_011704 | vanin 1 (Vnn1) | Detox | −0.60899 | x 0.656 | 0,0001 |
| NM_009676 | aldehyde oxidase 1 (Aox1) | Enz, | −0.60447 | x 0.658 | 0.0047 |
| NM_023785 | pro-platelet basic protein (Ppbp) | Signal, Immun | −0.59965 | x 0.66 | 0.0094 |
| NM_018778 | claudin 8 | Epm | −0.59480 | x 0.662 | 0.0003 |
| NM_009994 | cytochrome P450, 1b1, benz(a)anthracene inducible (Cyp1b1) | Enz, Detox | −0.59413 | x 0.662 | 0.0115 |
| NM_008165 | glutamate receptor, ionotropic, AMPA1 (alpha 1) (Gria1) | Recept | −0.59078 | x 0.664 | 0.0020 |
| NM_008592 | forkhead box C1 (Foxc1) | Trancript | −0.59037 | x 0.664 | 0.0382 |
| NM_007710 | creatine kinase, muscle (Ckmm) | Enz, MCm | −0.58630 | x 0.666 | 0.0162 |
| NM_007617 | caveolin 3 | Signal, MCm | −0.58227 | x 0.668 | 0.0273 |
| NM_007933 | enolase 3, beta muscle (Eno3) | Enz, MCm | −0.57902 | x 0.669 | 0.0328 |
| NM_009998 | cytochrome P450, 2b10, phenobarbitol inducible, type b (Cyp2b10) | Enz, Detox | −0.57377 | x 0.672 | 0.0312 |
| NM_008469 | keratin complex 1, acidic, gene 15 (Krt1-15) | Epm | −0.56978 | x 0.674 | 0.0318 |
| NM_008419 | potassium voltage-gated channel, member 5 (Kcna5) | Transport | −0.56587 | x 0.676 | 0.0363 |
| NM_007817 | cytochrome p450, 2f2 (Cyp2f2) | Enz, Detox | −0.56285 | x 0.677 | 0.0185 |
| NM_013494 | Carboxypeptidase E | Enz Prot metab | −0.56262 | x 0.677 | 0.0022 |
| NM_013712 | integrin beta 1 binding protein 2 | Recept, ECM | −0.56174 | x 0.677 | 0.0013 |
| NM_009814 | calsequestrin 2 (Casq2) | MCm | −0.55415 | x 0.681 | 0.0379 |
| BQ174827 | doublecortin and calciumcalmodulin-dependent protein kinase-like 1 | Enz, Prot metab | −0.55458 | x 0.68 | 0.0137 |
| NM_007429 | angiotensin II receptor, type 2 (Agtr2) | Recept | −0.55246 | x 0.682 | 0.0302 |
| NM_011098 | Brx1a mRNA for homeoprotein | Transcript, MCm | −0.54870 | x 0.684 | 0.0102 |
| AF356844 | carboxypeptidase Z | Enz, Prot metab | −0.54419 | x 0.685 | 0.0292 |
| NM_007751 | cytochrome c oxidase, subunit VIIIb (Cox8b) | Enz, Detox | −0.53763 | x 0.689 | 0.0133 |
| NM_011782 | a disintegrin-like and metalloprotease 5 (aggrecanase-2) (Adamts5) | Enz, ECM | −0.53751 | x 0.689 | 0.0316 |
| NM_023478 | uroplakin 3 (Upk3) | Epm | −0.53425 | x 0.69 | 0.0132 |
| NM_008664 | myomesin 2 | MCm | −0.53029 | x 0.692 | 0.0172 |
| NM_009610 | actin, gamma 2, smooth muscle, enteric (Actg2) | MCm | −0.52433 | x 0.695 | 0.0404 |
| NM_007868 | dystrophin, muscular dystrophy (Dmd) | MCm | −052267 | x 0.696 | 0.0067 |
| NM_080462. | histamine N-methyltransferase (Hnmt) | Enz, Immun | −0.52095 | x 0.697 | 0.0221 |
| NM_023617 | aldehyde oxidase 3 (Aox3) | Retino metab | −0.51687 | x 0.699 | 0.0094 |
| NM_025711 | asporin (Aspn) | ECM | −0.51470 | x 0.7 | 0.0146 |
| NM_007755 | cytoplasmic polyadenylation element binding protein (Cpeb) | Prot metab | −0.51347 | x 0.7 | 0.0004 |
| NM_017370 | haptoglobin (Hp) | Prot metab | −0.51060 | x 0.702 | 0.0175 |
| NM_008862 | protein kinase inhibitor, alpha (Pkia) | Prot metab | −0.50431 | x 0.705 | 0.0400 |
| NM_016762 | matrilin 2 | ECM | −050199 | x 0.706 | 0.0341 |
| NM_010390 | major histocompability complex Q1b | Immun | −0.50045 | x 0.707 | 0.0203 |
| NM_008242 | forkhead box D1 (Foxd1) | Transcript | −0.50023 | x 0.707 | 0.0268 |
| NM_008785 | serine (or cysteine) proteinase inhibitor, Clade A, member 5 (Serpina5) | Prot metab | −0.49974 | x 0.707 | 0.0366 |
| NM_009154 | semaphorin 5A (Sema5a) | Migr | −0.49740 | x 0.708 | 0.0409 |
| NM_020621 | pyrimidinergic receptor P2Y, G-protein coupled, 4 (P2ry4) | Recept | −049545 | x 0.709 | 0.0116 |
| M12233 | actin, alpha 1, skeletal muscle (Acta1) | MCm | −0.49534 | x 0.709 | 0.0005 |
| NM_008522.1 | lactotransferrin (Ltf) | Immun | −0.49078 | x 0,712 | 0.0160 |
| NM_053253.1 | Blu protein (Blu) | Immun | −0.48146 | x 0,716 | 0.0231 |
| NM_009144.1 | secreted frizzled-related sequence protein 2 (Sfrp2) | Prolif, EMT | −0.47671 | x 0,719 | 0.0211 |
| NM_011347.1 | selectin, platelet (Selp) | Recept, Adh | −0.47634 | x 0,719 | 0.0034 |
| NM_019662.1 | Ras-related associated with diabetes (Rrad) | Enz, GTPase | −0.47134 | x 0,721 | 0.0293 |
| NM_013669.1 | synaptosomal-associated protein, 91 kDa (Snap91) | Synapse marker | −0.46817 | x 0,723 | 0.0235 |
| NM_024406.1 | fatty acid binding protein 4, adipocyte (Fabp4) | Lipog | −0.46752 | x 0,723 | 0.0406 |
| AF357883.1 | homeodomain transcription factor (Nkx6-1) | Transcript | −0.46167 | x 0,726 | 0.0169 |
| AF020738.1 | fibroblast growth factor-related protein FGF-12A (Fgf12) | Sign | −0.45776 | x 0,728 | 0.0439 |
| NM_022881.1 | regulator of G-protein signaling 18 (Rgs18) | Signaling Regul | −0.45263 | x 0,731 | 0.0325 |
| NM_021467.2 | troponin I, skeletal, slow 1 (Tnni1) | MCm | −0.44982 | x 0,732 | 0.0163 |
| NM_019417.1 | PDZ and LIM domain 4 (Pdlim4) | Prot metab | −0.44649 | x 0,734 | 0.0225 |
| NM_010769.1 | matrilin 1, cartilage matrix protein 1 (Matn1) | ECM | −0.44476 | x 0,735 | 0.0280 |
| NM_019738.1 | nuclear protein 1 (Nupr1) | Transcript | −0.44316 | x 0,736 | 0.0051 |
| NM_023113.1 | aspartoacylase (aminoacylase) 2 | Enz, Prot metab | −0.43938 | x 0,737 | 0.0050 |
| NM_010669.1 | keratin complex 2, basic, gene 6b (Krt2-6b), | Epm | −0.43792 | x 0,738 | 0.0065 |
| NM_130895.1 | adenosine deaminase, RNA-specific, B1 (Adarb1) | Enz | −0.42746 | x 0,744 | 0.0152 |
| NM_031160.1 | ADP-ribosylation factor 4-like (Arf4l) | Signaling Regul | −0.42584 | x 0,744 | 0.0229 |
| NM_011582.1 | thrombospondin 4 (Thbs4) | ECM | −0.42420 | x 0,745 | 0.0242 |
| AF071068.1 | aromatic-L-amino-acid decarboxylase | Enz | −0.42030 | x 0,747 | 0.0194 |
| NM_010016.1 | decay accelerating factor 1 (Daf1) | Immun | −0.41955 | x 0,748 | 0.0062 |
| NM_011459.1 | serine protease inhibitor 8 (Spi8) | Enz | −0.41851 | x 0,748 | 0.0025 |
| NM_007940.1 | epoxide hydrolase 2, cytoplasmic (Ephx2) | Enz, Detox | −0.41824 | x 0,748 | 0.0242 |
| NM_007689.1 | chondroadherin (Chad) | ECM | −0.41822 | x 0,749 | 0.0327 |
Abbreviations: Funct. Cat., functional category of gene product; Adh, adhesion molecule; Detox, detoxification and xenobiotic metabolism; ECM, extracellular-matrix component or metabolism; Epm, epithelial marker; Enz, enzyme or enzyme inhibitor; Glycoconj, glycoconjugate metabolism; Immun, immune system; Inter metab, intermediary metabolism; Lipog, lipogenesis; MCm, muscle cell marker, Migr, cell migration; Prot metab, protein metabolism; Recept, receptor; Regul, regulator; Retino metab, retinoid metabolism, Signal, signaling molecule; Transcript, transcription factor or regulator; Transport, transporter molecule.
Genes with name and symbol in bold character had changes checked by RT-qPCR.
Data from RT-qPCR determinations used to confirm expression changes of selected genes and to determine their developmental expression profiles were referred to both 18S rRNA and Hprt1 mRNA. Although absolute values provided by both references were slightly different, they indicated the same proportional changes (Table 6). The use of either reference therefore did not affect conclusions. Because 18S rRNA level displayed constant proportionality with the amount of extracted RNAs and more stable 2−ΔΔct (i.e. more constant level) than Hprt1 mRNA across the developmental period under study, data presented below are those using 18S rRNA as reference.
Table 6.
Comparison of RT-qPCR data normalization by 18S rRNA and Hprt1 mRNA. Developmental changes of expression levels in the lung of 4 genes of interest (2 up-regulated and 2 down-regulated genes given as examples) are presented (arbitrary unit of 2−ΔΔct related to the expression of a calibrator sample).
| Adrenomedullin | Bone sialoprotein | C1qtnf3 | Crabp1 | |||||
|---|---|---|---|---|---|---|---|---|
| Reference gene | 18S | Hprt1 | 18S | Hprt1 | 18S | Hprt1 | 18S | Hprt1 |
| Day 0 | 0.51±0.11 | 0.72±0.13 | 0.25±0.07 | 0.40±0.15 | 0.61±0.18 | 1.10±0.50 | 0.02±0.00 | 0.03±0.01 |
| Day 4 | 0.13±0.03* | 0.20±0.01* | 1.22±0.09* | 2.33±0.67* | 1.99±0.42* | 3.42±0.46* | 0.08±0.02* | 0.14±0.03* |
| Day 7 | 0.50±0.16* | 0.59±0.02* | 1.04±0.26 | 1.33±0.19* | 1.10±0.11* | 1.55±0.21* | 0.46±0.03* | 0.67±0.12* |
| Day 10 | 0.75±0.30 | 1.10±0.26 | 0.66±0.22* | 1.07±0.29 | 0.85±0.16 | 1.72±0.46 | 0.71±0.16* | 1.27±0.15* |
| Day 14 | 0.49±0.13 | 1.52± 0.17 | 0.04±0.01* | 0.13±0.01* | 0.35±0.06* | 1.34±0.15 | 0.83±0.10 | 3.19±0.26* |
| Day 20 | 0.21±0.04* | 0.76±0.11* | 0.01±0.00* | 0.04±0.01* | 0.22±0.08 | 0.77±0.09* | 0.33±0.09* | 1.27±0.06* |
| Day 28 | 0.17±0.04 | 0.51±0.17 | 0.01±0.00 | 0.02±0.01 | 0.25±0.09 | 0.69±0.17 | 0.09±0.01* | 0.29±0.05* |
| Adult | 0.06±0.02* | 0.36±0.05 | 0.01±0.00 | 0.02±0.01 | 0.05±0.02* | 0.34±0.06* | 0.01±0.00* | 0.03±0.02* |
Mean ± se on 4 individual determinations per stage.
Significant difference from preceding stage (P<0.05 by U test).
Gene symbols: C1qtnf3 = C1q and tumor necrosis factor related protein 3; Crabp1: cellular retinoic acid binding protein 1.
Genes up regulated by FGF18 induction
To facilitate comparisons, the developmental profile of endogenous FGF18 is presented along with those of genes up regulated by FGF18-transgene induction (Fig. 4, bottom). FGF18 transcript was enhanced on P4 and further on P7 as compared with P0 level, then dropped on P14 and further on P28, and reached very low level in adult lung. Following transgene induction, the gene with most strongly increased expression (4-fold) was that of the vasoactive peptide adrenomedullin (Adm). RT-qPCR analysis on a larger number of pups indicated even stronger stimulation (almost 6.9-fold, table 7). Adm was followed by CD36 antigen (also known as fatty acid translocase), and hydroxysteroid 11β dehydrogenase 1 (Hsd11b1), which was confirmed by RT-qPCR although changes were found to be slightly (CD36) or largely (Hsd11b1) higher than those detected by microarrays (table 7). The postnatal expression profiles of these 3 genes in normal lung shared a 2.5- to 5-fold decrease between days P0 and P4 (Fig. 4), an event that appeared to have been prevented by FGF18 induction in transgenic pups. This was followed by re-increase from P4 but with different patterns. Adm transcript rose sharply from P4, peaked from P7 to P14, then fell to retrieve low level on P20 onwards (Fig. 4), displaying therefore a profile resembling that of endogenous FGF18 expression, except for the P4 decrease. CD36 expression also re-increased rapidly from P4 whereas that of Hsd11b1 re-increased more gradually, but contrary to Adm, both maintained high level until day 28 and returned to low level in adult lung only (Fig. 4). CD36 expression level varied widely among individuals.
Fig. 4.
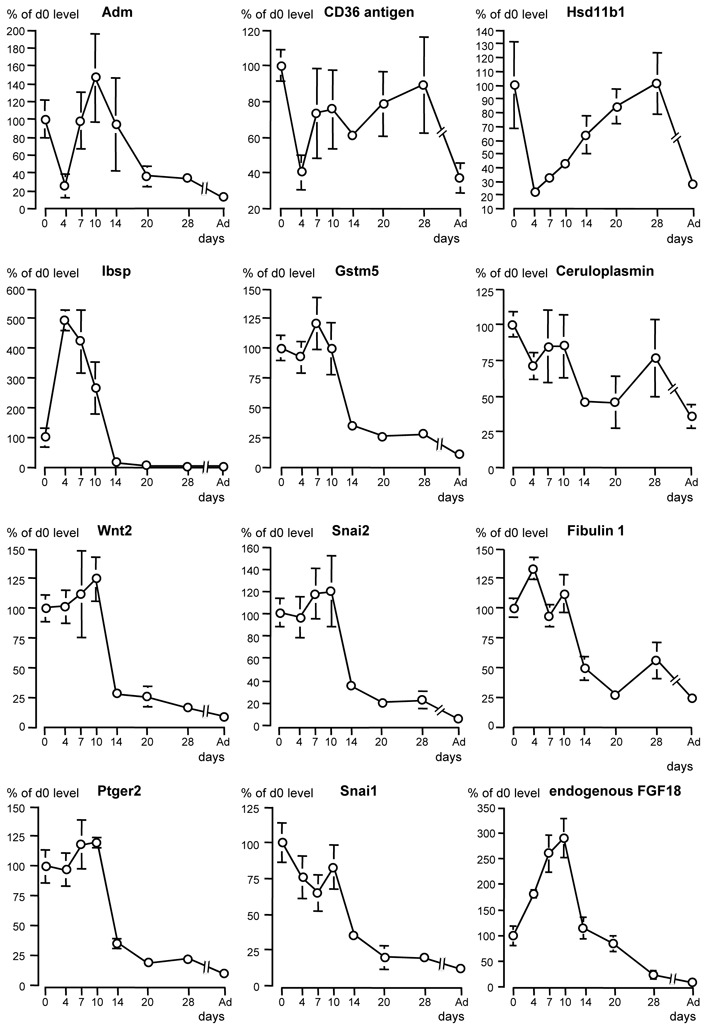
Developmental expression profile from birth to adulthood in wild-type mouse lung of 11 genes with expression found to be increased by FGF18-transgene induction in microarray analysis. Quantitative RT-qPCR was used to determine expression levels. Endogenous-FGF18 expression profile is also displayed for facilitating comparisons. Mean ± se on 4 individual lung samples from 2 different litters per stage (absence of error bar means that it was too small to be represented at this scale). Multiple stage comparison was made by Kruskal-Wallis analysis, followed by two-stage comparisons by Mann and Whitney U test. To avoid overloading of the graphs, significant differences are not represented, but significant changes are detailed in text. Gene symbols: Adm, adrenomedullin: Hsd11b1, hydroxysteroid 11β dehydrogenase 1; Gstm5, glutathione-S transferase, mu5; Wnt2, wingless-related MMTV integation site 2, Snai, snail homolog, Ptger2, prostaglandin R receptor EP2 subtype.
Table 7.
RT-qPCR determination of expression levels of 11 selected genes found to be up regulated by FGF18- transgene induction in microarray experiment.
| Genes | transcript level (2ΔΔCt) | % of mean control level per litter | ||
|---|---|---|---|---|
|
| ||||
| control pups | double-transgenic pups | control pups | double-transgenic pups | |
|
|
||||
| Angiogenesis | ||||
| Adrenomedullin | 2.16 ± 0.26 | 14.23 ± 2.59*** | 100.0 ± 9.6 | 686.1 ± 136.1*** |
| Wnt2 | 2.47 ± 0.32 | 5.54 ± 0.74*** | 100.0 ± 7.5 | 242.5 ± 39.4** |
|
| ||||
| ECM components | ||||
| Bone sialoprotein | 1.95 ± 0.20 | 4.99 ± 0.70*** | 100.0 ± 7.7 | 281.4 ± 61.6** |
| Fibulin 1 | 1.83 ± 0.23 | 2.91 ± 0.23* | 100.0 ± 6.9 | 168.6 ± 14.7** |
|
| ||||
| Cell migration | ||||
| Snai1 | 2.54 ± 0.38 | 3.97 ± 0.42* | 100.0 ± 8.1 | 161.4 ± 16.2** |
| Snai2 | 2.42 ± 0.31 | 5.51 ± 1.21* | 100.0 ± 8.1 | 242.4 ± 59.8* |
| Ptger2 | 1.31 ± 0.12 | 2.31 ± 0.26** | 100.0 ± 7.0 | 176.5 ± 20.7** |
|
| ||||
| Antioxidant mech. | ||||
| Gstm5 | 1.69 ± 0.21 | 5.09 ± 0.86*** | 100.0 ± 7.7 | 321.43 ± 64.2** |
| Ceruloplasmin | 1.96 ± 0.38 | 4.54 ±0.66** | 100.0 ± 12.3 | 263.17 ± 52.7** |
|
| ||||
| Other | ||||
| CD36 antigen | 2.37 ± 0.42 | 4.97 ± 0.74** | 100.0 ± 10.7 | 250.8 ± 38.0** |
| Hsd11b1 | 1.33 ± 0.14 | 6.10 ± 1.17*** | 100.0 ± 8.5 | 491.0 ± 102.1*** |
Mean ± se on 10 control pups and 11 double-transgenic pups from 5 different litters. Difference with control group (one-tailed t test for unpaired data) for:
P<0.05;
P<0.01;
P<0.001.
Genes are presented in decreasing order of stimulation gained from microarray analysis. Gene symbols: Hsd11b1 = hydroxysteroid 11-beta dehydrogenase 1; Gstm5 = glutathione S-transferase, mu5; Wnt2 = wingless-related MMTV integration site 2; Snai = snail homolog; PgEPR EP2 = prostaglandin E receptor, EP2 subtype.
In addition to fibulin 1 expression already known to be enhanced by FGF18 (7), the expression of various extracellular matrix (ECM) components was increased. These include bone sialoprotein (or integrin binding sialoprotein, Ibsp), chondroitin sulfate proteoglycan 2 (table 4), and chondromodulin-1b (not included in table 4). RT-qPCR confirmed bone sialoprotein and fibulin 1 changes, the steady-state level of their transcripts being increased 2.8 times and 1.69 times, respectively (table 6). Their developmental profiles presented high level during the period of secondary septation and a drop on day 14 (Fig. 4), consistent with control by endogenous FGF18.
FGF18 induction up-regulated the expression of a variety of genes involved in antioxidant metabolism, including glutathione-S-transferase mu5 (Gstm5), ceruloplasmin, leucine-rich α2-glycoprotein-1 (Lrg1) cytochrome P450 (Cyp) 2e1, cysteine dioxygenase 1, adrenodoxin (table 4), serine hydroxymethyl transferase, carbonyl reductase 1, and SH3-binding domain glutamic acid-rich protein like (Sh3bgrl) (not included in table 4). Again, expression changes determined by RT-qPCR for the mostly affected genes Gstm5 and ceruloplasmin were larger than those from microarrays, reaching 3.2-fold and 2.6-fold, respectively (table 7). Gstm5 developmental expression profile displayed increase from P4 to P7, fall on day 14, and further decrease to very low level in adulthood, consistent with control by endogenous FGF18 (Fig. 4). By contrast, that of ceruloplasmin displayed postnatal decrease, no significant change from P4 to P10, moderate decrease on P14, increase on P28, and maintenance of sizeable level in adult lung, which argues for multifactorial control involving factors other than FGF18.
FGF18 induction also more or less enhanced (+50% to +16%) the expression of genes the protein products of which are known to stimulate or are likely to be involved in cell migration, including wingless-related MMTV integration site 2 (Wnt2), snail homologs 1 and 2 (Snai1, Snai2), sushi-repeat protein, mixed-lineage kinase-like mitogen-activated triple kinase alpha (MLTKα), and neural precursor cell-expressed developmentally down-regulated gene 9 (Nedd9). Importantly, it simultaneously stimulated (+100% to +16.5%) the expression of genes the products of which have opposite and antifibrotic effects, including (in addition to CD36 antigen already mentioned) Th2-specific cytokine FISP/IL24, Borg4/binder of Rho GTPase 4, prostanoid E receptor EP2 subtype (Ptger2) (table 4), and the tumor suppressor Lim domains-containing protein 1 (Limd1, not included in table 4). RT-qPCR analysis run for Wnt2, Snai1, Snai2, and Ptger2 confirmed enhancement with larger changes than those detected by microarray analysis (table 7), particularly marked for Wnt2 and Snai2 (2.4-fold each). Developmental expression profiles of these 4 genes were very similar, with high level during alveolar septation, maximum on P7-10, abrupt drop on P14, and low level during alveolar-wall maturation and in adult lung (Fig. 4), consistent with regulation by FGF18.
Last, in keeping with its effects on lung-fibroblast proliferation (7), FGF18 enhanced the expression of a variety of genes involved in cell growth and ribosome synthesis, including G0G1 switch gene 2, brix domain containing 1 (Bxdc1), NIMA-related expressed kinase 8 (Nek8) (table 4), and more slightly (not included in table 4), mitogen activated protein kinase kinase 1 (Map2k1), mitogen activated protein kinase kinase kinase 8 (Map3k8), cyclin D2, ribosomal proteins S2 and S13, mitochondrial-ribosomal protein S18B, and WD repeat domain 12 (Wdr12).
Genes down regulated by FGF18 induction
Six genes presenting over 50% decreased expression (table 5) included the adipocyte markers (also expressed in alveolar type II cells) C1q and tumor necrosis factor related protein 3 (C1qtnf3 or CTRP3, also known as cartducin, cartonectin, or collagenous repeat-containing sequence of 26kdA protein [CORS 26]), stearoyl-Coenzyme A desaturase 1 (Scd1), and Ca(2+)-sensitive chloride channel 2, the alveolar type II cell marker ATP-binding cassette transporter 3 (ABCA3), the ECM component osteoglycin (Ogn), and the ECM-degrading enzyme, fibroblast activation protein (Fap) also designated Seprase. Other ECM and connective tissue components, including proteoglycan 4, chondrolectin, asporin, matrilins 1 and 2, thrombospondin 4 and chondroadherin, and another ECM-degrading enzyme, Adamts 5, had notably reduced expression (−47% to −30%, table 5). The transcript of the transcription factor high mobility group box protein/SRY-box containing gene (Sox2), another gene with epithelium-restricted expression, was reduced by 40%. RT-qPCR analysis confirmed the decrease for C1qtnf3, Scd1, ABCA3 (−49%, −69%, −48%, respectively), but not for Sox 2, Ogn and Fap (table 8). As regards Sox 2, Ogn and Fap, although RT-qPCR confirmed lower levels in those samples used for microarray hybridization, the variability between and inside litters was so wide for single- as well as for double-transgenic pups that no significant change was observed on the average in the 11 double-transgenic pups compared to their 10 control littermates. Moreover, there was no correlation between individual values and the extent of FGF18 changes in double-transgenic pups, suggesting the fortuitous character of variations. Developmental expression profiles were established for C1qtnf3, Scd1 and ABCA3 (Fig. 5). C1qtnf3 transcript rose from P0 to P4, which was unlikely to be related to endogenous FGF18, decreased then gradually until P14, which could relate to FGF18 peak, and decreased further until adulthood. Scd1 transcript increased modestly from P4 to P14, then strongly until P28 to decline in adult lung. ABCA3 transcript declined half on P4, rose about twice on days 7–10, which could indeed not be due to endogenous FGF18, then decreased about twice on P14 and remained stable thereafter.
Table 8.
RT-qPCR determination of expression levels of 10 selected genes found to be down regulated by FGF18-transgene induction in microarray experiment.
| Genes | transcript level (2ΔΔCt) | % of mean control level per litter | ||
|---|---|---|---|---|
|
| ||||
| control pups | double-transgenic pups | control pups | double-transgenic pups | |
| Adipocyte and/or alveolar typeII-cell markers | ||||
| C1qtnf3 | 1.10 ± 0.16 | 0.62 ± 0.19* | 100.0 ± 8.6 | 50.9 ± 12.7** |
| Stearoyl-CoA desaturase 1 | 1.83 ± 0.24 | 0.57 ± 0.06*** | 100.0 ± 9.7 | 30.7 ± 3.4*** |
| Abca3 | 1.52 ± 0.49 | 0.84 ± 0.32*** | 100.0 ± 20.9 | 54.8 ± 15.6*** |
|
| ||||
| Other epithelial-cell markers | ||||
| Cyp1b1 | 1.31 ± 0.18 | 0.96 ± 0.12* | 100.0 ± 7.6 | 76.1 ± 9.6* |
| Sox2 | 1.24 ± 0.20 | 1.29 ± 0.14 ns | 100.0 ± 7.9 | 93.4 ± 13.7 ns |
|
| ||||
| ECM and ECM metabolism | ||||
| Osteoglycin | 1.92 ± 0.33 | 1.70 ± 0.43ns | 100.0 ± 10.8 | 94.2 ± 24.1ns |
| Fap/seprase | 1.60 ± 0.90 | 1.51 ± 0.99ns | 100.0 ± 8.1 | 104.4 ± 28.0ns |
|
| ||||
| Cell migration/invasion | ||||
| Doublecortin | 1.80 ± 0.20 | 1.13 ± 0.28* | 100.00 ± 9.89 | 63.0 ± 16.9* |
|
| ||||
| Retinoic acid metabolism | ||||
| Crabp1 | 1.74 ± 0.17 | 0.72 ± 0.26** | 100.0 ± 7.5 | 41.4 ± 19.9** |
| Aldh1a1 | 1.83 ± 0.18 | 2.29 ± 0.45 ns | 100.0 ± 8.1 | 133.1 ± 27.9 ns |
Mean ± se on 10 control pups and 11 double-transgenic pups from 5 different litters. Difference with control group (one-tailed t test for unpaired data) for:
P<0.05;
P<0.01;
P<0.001;
ns = not significant. Genes are presented in decreasing order of inhibition gained from microarray analysis. Gene symbols and abbreviation: C1qtnf3 = C1q and tumor necrosis factor related protein 3; CoA = coenzyme A; Abca3 = ATP-binding cassette transporter ABCA3; Fap = fibroblast activation protein; Crabp1: cellular retinoic acid binding protein 1; Sox2 = SRY-box containing gene 2; Aldh1a1 = aldehyde dehydrogenase 1, subfamily A1; Cyp1b1 = cytochrome P450, 1b1.
Fig. 5.
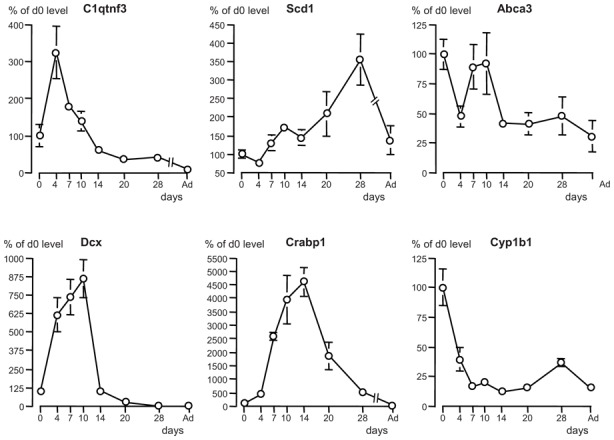
Developmental expression profile from birth to adulthood in wild-type mouse lung of 6 genes with expression found to be decreased by FGF18-transgene induction in microarray analysis. Quantitative RT-qPCR was used to determine expression levels. Mean ± se on 4 individual lung samples from 2 different litters per stage (absence of error bar means that it was too small to be represented at this scale). Multiple stage comparison was made by Kruskal-Wallis analysis, followed by two-stage comparisons by Mann and Whitney U test. To avoid overloading of the graphs, significant differences are not represented, but significant changes are detailed in text. Gene symbols: C1qtnf3, C1q and tumor necrosis factor related protein 3; Scd1, stearoyl-coenzyme A desaturase 1; Abca3, ATP-binding cassette transporter ABCA3; Dcx, doublecortin, Crabp1, cellular retinoic acid binding protein 1; Cyp1b1, cytochrome P450, 1b1.
Two genes whose protein products are related to retinoic acid (RA) metabolism, cellular RA binding protein 1 (Crabp1), aldehyde dehydrogenase family 1, subfamily A1 (Aldh1a1) displayed reduced expression in microarray findings (−45% and −40%, respectively). This was confirmed by RT-qPCR for Crabp1 (−59%) but not for Aldh1a1 for the same reasons as above (table 8). However, Crabp1 developmental profile presented huge increase from P4 to P14 and decreased then progressively to extremely low level in adult lung (Fig. 5). This profile is not consistent with a negative control by endogenous FGF18 since both transcripts evolve in parallel during development. FGF18 induction reduced to variable extent the expression of various genes related with cell migration and invasion, including doublecortin (Dcx, −48%), plasminogen activator inhibitor type II (PAI2/Serpinb2, −39%), cytoplasmic polyadenylation element binding protein (Cpeb, −30%), and semaphorin 5A (−29%) (table 5), and more modestly PAI1/Serpine 1 (−23%, not included in table 5). RT-qPCR analysis of Dcx confirmed decrease (table 7). As for Crabp1, Dcx developmental expression profile looked similar to that of FGF18 with peak from P7 to P10 and fall on P14 (Fig. 5), which indicates that endogenous FGF18 does not exert negative control.
In addition to ABCA3, FGF18 induction reduced the expression of a variety of alveolar and/or bronchiolar (Clara) epithelial cell markers, including cytochrome oxidases Cyp1b1 (confirmed by RT-qPCR, table 7), Cyp2f2 and Cyp2b10, aquaporins 3 and 4, uteroglobin-related protein 1A/secretoglobin (Scgb3a2), aldehyde oxydase 3, and claudin 8. Expressions of other genes related with oxidant/detoxification/xenobiotic metabolism, namely Cyp2b20, Cyp4b20, Cyp2f2, cytochrome oxydase subunits VIIa and VIIIb, vanin1 (table 5), and Cyp2b20 (−21%, not included in table 5), were also diminished. The developmental expression profile of Cyp1b1 was consistent with down regulation by endogenous FGF18: initially elevated at birth, it decreased abruptly to minimum level maintained from d7 to d20, re-increased slightly on d28, and decreased again in adult lung (Fig. 5).
Last, FGF18 induction decreased the expression of a variety of contractile cell markers, including troponins C, I and T3, myomesin 1, chondrolectin, actinin alpha 2, mouse alpha cardiac myosin heavy chain, calponin 1, myomesin 2, actins gamma 2 and alpha 1, dystrophin, caveolin 3, and calsequestrin 2 (−45% to −25%, table 5). Interestingly, expression of the specific myofibroblast markers calponin 1 (table 5) and α-smooth-muscle actin (more slightly diminished, not shown in table 5) was also reduced.
DISCUSSION
Using a mouse transgenic model, we found that advancing the peak of pulmonary FGF18 expression affected the expression level of numerous genes in a variety of pathways. Despite apparent complexity, most of these effects can be filed into 3 main groups that deal with (i) angiogenesis, (ii) cell migration, and (iii) expression of ECM components. Although alveolarization was not advanced by FGF18 induction, these effects are suggestive of FGF18 involvement in alveolarization mechanisms.
Putative FGF18 target cells in the lung considering FGF18 receptor expression
FGF18 binds with high affinity the IIIc splice variant of FGFR3 and the unique form of FGFR4, and more modestly the IIIc splice variant of FGFR2, but presents no affinity for IIIb isoforms (12, 39). Considering FGFR expression pattern, all cell types of lung parenchyma are potential targets of FGF18 through one or two receptors. Thus, if FGFR2 IIIc variants are expressed exclusively in mesenchyme-derived tissues, both lung mesenchymal and alveolar epithelial cells express FGFR3 and FGFR4 (41), although it is unknown whether they express differentially FGFR3IIIb and IIIc. Pulmonary endothelial cells and vascular smooth muscle cells are likely to express FGFR2IIIc and FGFR3IIIc, since these have been characterized in human umbilical vascular endothelial cells (HUVEC) and aortic smooth muscle cells (2). Indeed, direct although different biological effects of FGF18 have been demonstrated on cultured cells of the different types. Specifically, FGF18 enhanced lung fibroblast proliferation and let unchanged epithelial-cell proliferation (7), whereas it induced BMP4 expression in tracheal epithelial cells (14). HUVEC displayed chemotaxis toward FGF18, but failed to proliferate in response to the factor (1). Studying changes in gene expression level in whole lung therefore allows one to detect changes occurring in any of potential target cells. However, this prevents from identifying any compartmentalization influence, except for genes known to present expression restricted to one cell type. The present study should therefore be completed by future investigations aiming at spatial determination of FGF18 effects.
Limitations of the study
One could raise an objection that lack of morphological changes after precocious induction of FGF18 expression argues against the involvement of the factor in alveolarization mechanisms. However, considering the complexity of alveolarization and the multiplicity of involved control factors (5) it is not surprising that the induction of a single factor, even if actually involved, was insufficient alone to induce precociously the whole process. More questionable are apparently paradoxical effects of transgenic FGF18 such as markedly increased CD36 and Hsd11b1 expressions. Although endogenous FGF18 might contribute to their expression re-increase from P4, their profiles, contrary for instance to that of Adm, do not coincide with that of FGF18. Hsd11b1 expression remains at low relative level during septation when FGF18 peaks. CD36 expression remains elevated and that of Hsd11b1continues increasing when FGF18 expression falls to maintain maximal level until P28, i.e. 14 days after FGF18 fall down. This appears inconsistent with control by FGF18 only, and presumably results from complex changes in the balance of various antagonistic control mechanisms.
Nonetheless, the herein described maintenance of Hsd11b1 expression at levels much lower than P0 level during septation period is novel and developmentally highly significant. Although in cell-free system, this interconverting enzyme of cortisone and cortisol behaves mainly as a dehydrogenase (inactivation), in vivo it is considered to perform 11β-reduction (activation) (10). Because corticosteroids have been reported to inhibit septation and to terminate alveolarization through fusion of the double capillary layer and thinning of septa (32, 43), low pulmonary corticosteroid activation subsequent to low Hsd11b1 expression level therefore appears as requisite for septation. Pursued elevation of Hsd11b1 expression between days 14 and 28 appears conversely to relate to corticosteroid-driven septation arrest and maturation of septa.
Several issues could account for difficulty in interpreting FGF18-induced effects. In the lung, FGF18 is released by interstitial cells (3, 7, 51), and possibly endothelial cells (2). The use of SP-C regulatory sequence to target FGF18 transgene expression on alveolar epithelial type II cells therefore leads to ectopic expression. This might have changed local availability of the factor, and it cannot be ruled out that this might have affected the responsiveness of its target cells, which could have been somehow different from that to FGF18 expressed from endogenous gene. Also likely to be involved in possible differences appear changes in cellular microenvironment, i.e. interaction of FGF18 with signal molecules either different or present in different relative proportions in the pre- and postnatal lung. Hence, paradoxical changes observed in double-transgenic pups might have been permitted by conditions prevailing around birth that were different from those during alveolarization. In addition, difficulties could be inherent to the model itself because of possible toxicity of the SP-C-rtTA transgene and because doxycycline is a pan-MMP (matrix metalloproteinase) inhibitor while MMPs play a functional role in alveolarization (discussed in 52). The absence of lung morphological changes in developing (this study) or adult (51) mice of this strain however argues against this assumption. Last, RT-qPCR post-hoc analysis gave more consistent results for up-regulated than for down-regulated genes. This appeared to be due to wide random expression variations of genes that presented by chance lower expression in those 4 double-transgenic lung samples used for microarray analysis. Because it was indeed not possible to perform individual RT-qPCR control for all gene expressions found to be diminished in microarray data, decreases were considered to be effective when there was consistency for several genes among a group (e.g. cell-type markers or genes encoding products with similar functions or involved in a same pathway). These considerations point the interest of examining the developmental expression profile of putative target genes to interpret results from transcriptome analysis.
Adrenomedullin and genes involved in angiogenesis
The primary target of FGF18 was Adm gene, the expression of which was increased about 7-fold by transgene induction. Adm developmental expression profile, which, similar to that of FGF18 displayed peak coincidental with secondary septation, was consistent with control by endogenous FGF18. Adrenomedullin is hypoxia-inducible peptide with vasodilator, antioxidative and angiogenic properties, highly expressed in the developing lung. Not only has Adm been reported to regulate cell growth and survival, to induce proliferation and migration of endothelial progenitor cells, to enhance the regenerating potential of the latter for pulmonary endothelium (36), and to regenerate alveoli and vasculature in elastase-induced pulmonary emphysema (35), but its role in alveolar development was recently evidenced (46). Lung expression of its transcript increases during alveolar development in rat lung also, and Adm requirement for alveolar septation and angiogenesis was demonstrated in this species (46). Whereas intranasal administration of an adrenomedullin-antagonist decreased lung VEGF expression and capillary density, and impaired alveolar development, giving exogenous adrenomedullin in a hyperoxic model of BPD reciprocally attenuated arrested lung angiogenesis and alveolar development (46).
FGF18-transgene induction also enhanced expression of other genes likely to be involved in angiogenesis, including Wnt2, a growth and differentiation factor for endothelial cells (22). This is consistent with decreased Wnt2 expression reported in compound FGFR3/4 mutant mice (42). Although more modestly, FGF18 also enhanced the expression of cullin 2 (not shown in table 4), which is required for vascular development through regulation of VEGF transcription mediated by hypoxia-inducible factor α (29).
The importance of FGF18 for angiogenesis had been evidenced previously in other organs by its chemotactic effect for endothelial cells (1) and by delayed skeletal vascularization in FGF18−/− mice (27). Consistently, vessel size and PECAM labeling were increased in the lung of SP-C-rtTA/(teto)7-FGF18 mouse fetuses treated with doxycycline during whole gestation (51). Our findings now identify FGF18 as a likely important player in the control of alveolar angiogenesis, an event that is absolute requirement for alveolarization and is compromised in BPD.
FGF18 effects related with cell migration
FGF18 is known to drive cell migration, especially for endothelial cells (1, 27), which is in keeping with effects of FGF18 transgene on a panel of genes involved in this process. In addition to Adm, these include the zinc-finger transcription factor-encoding genes Snai1 (Snail) and Snai2 (Slug), sushi repeat protein, Lrps 1 and 2, and Nedd 9. However, in addition to promotion of cell movement during organogenesis, SNAI proteins are also well known to be involved in epithelial-mesenchymal transition (EMT) and metastatic invasion (37). In the lung, their overexpression was sufficient to induce EMT in alveolar epithelial cells whereas SNAI depletion conversely attenuated TGFβ1-induced cell migration and EMT (18). Up regulation of Wnt2 and down regulation of Sfrp1, both induced by FGF18, are also involved in EMT (20). Although these FGF18 effects could therefore appear as favoring EMT, there is no EMT during normal alveolar development. EMT is by contrast encountered in pathological condition, namely lung fibrosis, via activation of endogenous latent TGFβ1 (21). Among other possible mechanisms, prevention of EMT during alveolarization could be due to the simultaneous induction of counteracting mechanisms by FGF18, including (i) down-regulation of genes encoding factors involved in cell invasion and metastasis such as doublecortin (13), PAI1 and PAI2 (33), and semaphorin 5A (40), and (ii) up-regulation of genes that inhibit spreading of mesenchyme-derived cells including fibulin-1 (53) and FISP/IL24 (9).
Also consistent appears the FGF18-induced reduced expression of numerous muscle-cell markers, which are expressed in the lung by smooth muscle cells and myofibroblasts. FGF18 thus reduced notably the expression of genes that are known to be down-regulated at initiation of mouse lung alveolarization (54), but were conversely up-regulated in TGFβ1-induced fibroblast-to-myofibroblast transdifferentiation (44) or in experimental hyperoxia-induced BPD (47). Prostaglandin E2 has been repeatedly reported to modulate lung inflammation and to prevent the fibroblast-to-myofibroblast transition through E prostanoid receptor 2 (Ptger2) signal (23, 50). Taken together, the increased number of pulmonary myofibroblasts in compound FGFR3/4 mutant mice (42) and the increased expression of Ptger2 consecutive to FGF18 induction evidenced herein strongly argue for a physiological role of FGF18 in prevention of fibrosis. Although myofibroblast differentiation and migration are necessary to alveolarization (26), FGF18 might prevent these from becoming excessive and falling over into EMT and fibrosis.
Effects on ECM components
FGF18 strongly enhanced the expression of the ECM components bone sialoprotein, chondroitin sulfate proteoglycans, and fibulin 1. Fibulin 1 is clearly indispensable to normal alveolar development (24). The role of bone sialoprotein in alveolarization is unknown, but its expression peak coincidental with secondary septation and its angiogenic properties (38) argue for involvement. As regards chondroitin sulfate proteoglycans, they are known to be present in alveolar basement membrane. FGF18 conversely decreased the expression of a variety of other ECM components. Previous investigations (4, 31, 54) have evidenced clusters of ECM-component genes up regulated either early during the phase of secondary septation or later during maturation of septa. FGF18 appears as an inducer of genes belonging to the former group. Interestingly, the expression of the ECM-related genes asporin and microfibrillar protein 5 that was increased in FGFR3/4-null mice (42) was conversely decreased by FGF18 induction, suggesting that FGF18 decreases expression of the late-group genes.
Other effects
FGF18 enhanced the expression of a variety of cytochrome oxidases and of genes involved in antioxidant mechanisms. The postnatal developmental expression profiles of glutathione-S-transferase and ceruloplasmin, with high postnatal level and fall on day 14, were consistent with control by FGF18. This is interesting in a therapeutic perspective, because preterm infants have immature antioxidant defense mechanisms and increased susceptibility to oxidative stress, which in turn is precipitating factor of BPD. Conversely, FGF18 down regulated a number of P450 cytochrome oxidases and other genes involved in xenobiotic metabolism. Numerous cytochrome oxidases with FGF18-decreased expression, including Cyp2b10, Cyp2b20, Cyp2f2 and Cyp4b1 were recently found to be down regulated in the alveolarizing postnatal lung (54). It was suggested that these changes could be related to expansion of epithelial precursor cell pool through balance between proliferation and differentiation of epithelial cells (54). Consistent with this assumption is the FGF18-diminished expression of a number of genes expressed selectively in lung epithelial cell subsets, including Ca(2+)-sensitive chloride channel 2, Abca3, Cyp1b1, Cyp2f2, stearoyl coenzyme A dehydrogenase (Scd1), aquaporins 3 and 4, uteroglobin-related protein 1a, and aldehyde oxidase 3. However, targeting the expression of FGF18 transgene on type II cells by the use of SP-C promoter might have enhanced these effects.
Lastly, it is worth pointing that, although not consistent with a negative control by endogenous FGF18, the developmental expression profile of Crabp1 remarkably presents a peak coincidental with alveolar septation. Taking into account the central role of RA released by lipofibroblasts in alveolarization (5), this appears to correspond to maximal capacity of lung cells to link and retain RA.
Acknowledgments
SP-C-rtTA-FGF-18 founder mice were obtained from the Cincinnati Children’s Research Foundation through material transfer agreement established on October 28, 2003.
References
- 1.Antoine M, Wirz W, Tag CG, Gressner AM, Wycislo M, Muller R, Kiefer P. Fibroblast growth factor 16 and 18 are expressed in human cardiovascular tissues and induce on endothelial cells migration but not proliferation. Biochem Biophys Res Commun. 2006;346:224–233. doi: 10.1016/j.bbrc.2006.05.105. [DOI] [PubMed] [Google Scholar]
- 2.Antoine M, Wirz W, Tag CG, Mavituna M, Emans N, Korff T, Stoldt V, Gressner AM, Kiefer P. Expression pattern of fibroblast growth factors (FGFs), their receptors and antagonists in primary endothelial cells and vascular smooth muscle cells. Growth Factors. 2005;23:87–95. doi: 10.1080/08977190500096004. [DOI] [PubMed] [Google Scholar]
- 3.Boucherat O, Benachi A, Barlier-Mur AM, Franco-Montoya ML, Martinovic J, Thebaud B, Chailley-Heu B, Bourbon JR. Decreased lung fibroblast growth factor 18 and elastin in human congenital diaphragmatic hernia and animal models. Am J Respir Crit Care Med. 2007;175:1066–1077. doi: 10.1164/rccm.200601-050OC. [DOI] [PubMed] [Google Scholar]
- 4.Boucherat O, Franco-Montoya ML, Thibault C, Incitti R, Chailley-Heu B, Delacourt C, Bourbon JR. Gene expression profiling in lung fibroblasts reveals new players in alveolarization. Physiol Genomics. 2007;32:128–141. doi: 10.1152/physiolgenomics.00108.2007. [DOI] [PubMed] [Google Scholar]
- 5.Bourbon J, Delacourt C, Boucherat O. Basic mechanisms of alveolarization. In: Abman SH, editor. Bronchopulmonary dysplasia New York: Informa Healthcare. 2009. pp. 56–88. [Google Scholar]
- 6.Bourbon JR, Boucherat O, Boczkowski J, Crestani B, Delacourt C. Bronchopulmonary dysplasia and emphysema: in search of common therapeutic targets. Trends Mol Med. 2009;15:169–179. doi: 10.1016/j.molmed.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Chailley-Heu B, Boucherat O, Barlier-Mur AM, Bourbon JR. FGF-18 is upregulated in the postnatal rat lung and enhances elastogenesis in myofibroblasts. Am J Physiol Lung Cell Mol Physiol. 2005;288:L43–51. doi: 10.1152/ajplung.00096.2004. [DOI] [PubMed] [Google Scholar]
- 8.Chailley-Heu B, Boucherat O, Benachi A, Bourbon JR. FGF18 is up-regulated at the onset of alveolar septation in rodent and human lungs. Proc Am Thor Soc. 2006;3:A674. [Google Scholar]
- 9.Chen J, Chada S, Mhashilkar A, Miano JM. Tumor suppressor MDA-7/IL-24 selectively inhibits vascular smooth muscle cell growth and migration. Mol Ther. 2003;8:220–229. doi: 10.1016/s1525-0016(03)00176-x. [DOI] [PubMed] [Google Scholar]
- 10.Draper N, Stewart PM. 11beta-hydroxysteroid dehydrogenase and the pre-receptor regulation of corticosteroid hormone action. J Endocrinol. 2005;186:251–271. doi: 10.1677/joe.1.06019. [DOI] [PubMed] [Google Scholar]
- 11.Elluru RG, Thompson F, Reece A. Fibroblast growth factor 18 gives growth and directional cues to airway cartilage. Laryngoscope. 2009;119:1153–1165. doi: 10.1002/lary.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Walsh CA. Protein-protein interactions, cytoskeletal regulation and neuronal migration. Nat Rev Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 14.Hyatt BA, Shangguan X, Shannon JM. BMP4 modulates fibroblast growth factor-mediated induction of proximal and distal lung differentiation in mouse embryonic tracheal epithelium in mesenchyme-free culture. Dev Dyn. 2002;225:153–165. doi: 10.1002/dvdy.10145. [DOI] [PubMed] [Google Scholar]
- 15.Ihaka R, Gentleman R. R: a language for data analysis and graphics. Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 16.Irizarry RA, Gautier L, Cope L. An R package for analysis of Affymetrix oligonucleotide arrays. In: Parmigiani G, Garrett ES, Irizarry SA, Zeger SL, editors. The analysis of Gene Expression Data: Methods and Software: Methods and software. New York: Springer; 2002. [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Jayachandran A, Konigshoff M, Yu H, Rupniewska E, Hecker M, Klepetko W, Seeger W, Eickelberg O. SNAI transcription factors mediate epithelial-mesenchymal transition in lung fibrosis. Thorax. 2009;64:1053–1061. doi: 10.1136/thx.2009.121798. [DOI] [PubMed] [Google Scholar]
- 19.Jobe AH, Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;163:1723–1729. doi: 10.1164/ajrccm.163.7.2011060. [DOI] [PubMed] [Google Scholar]
- 20.Katoh M. Epithelial-mesenchymal transition in gastric cancer (Review) Int J Oncol. 2005;27:1677–1683. [PubMed] [Google Scholar]
- 21.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103:13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein D, Demory A, Peyre F, Kroll J, Augustin HG, Helfrich W, Kzhyshkowska J, Schledzewski K, Arnold B, Goerdt S. Wnt2 acts as a cell type-specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology. 2008;47:1018–1031. doi: 10.1002/hep.22084. [DOI] [PubMed] [Google Scholar]
- 23.Kolodsick JE, Peters-Golden M, Larios J, Toews GB, Thannickal VJ, Moore BB. Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am J Respir Cell Mol Biol. 2003;29:537–544. doi: 10.1165/rcmb.2002-0243OC. [DOI] [PubMed] [Google Scholar]
- 24.Kostka G, Giltay R, Bloch W, Addicks K, Timpl R, Fassler R, Chu ML. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–7034. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindahl P, Karlsson R, Hellstrom M, Gebre-Medhin S, Willets K, Heath JK, Betsholtz C. Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development. 1997;124:3943–3953. doi: 10.1242/dev.124.20.3943. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 28.Lopez E, Boucherat O, Franco-Montoya ML, Bourbon JR, Delacourt C, Jarreau PH. Nitric oxide donor restores lung growth factor and receptor expression in hyperoxia-exposed rat pups. Am J Respir Cell Mol Biol. 2006;34:738–745. doi: 10.1165/rcmb.2005-0254OC. [DOI] [PubMed] [Google Scholar]
- 29.Maeda Y, Suzuki T, Pan X, Chen G, Pan S, Bartman T, Whitsett JA. CUL2 is required for the activity of hypoxia-inducible factor and vasculogenesis. J Biol Chem. 2008;283:16084–16092. doi: 10.1074/jbc.M710223200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariani T. Regulation of alveogenesis by reciprocal proximodistal fibroblast growth factor and retinoic acid signaling. Am J Respir Cell Mol Biol. 2004;31:S52–S57. [Google Scholar]
- 31.Mariani TJ, Reed JJ, Shapiro SD. Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol. 2002;26:541–548. doi: 10.1165/ajrcmb.26.5.2001-00080c. [DOI] [PubMed] [Google Scholar]
- 32.Massaro D, Massaro GD. Dexamethasone accelarates postnatal alveolar wall thinning and alters wall composition. Am J Physiol Regul Integr Comp Physiol. 1986;251:R218–R224. doi: 10.1152/ajpregu.1986.251.2.R218. [DOI] [PubMed] [Google Scholar]
- 33.McMahon B, Kwaan HC. The plasminogen activator system and cancer. Pathophysiol Haemost Thromb. 2008;36:184–194. doi: 10.1159/000175156. [DOI] [PubMed] [Google Scholar]
- 34.Mesas-Burgos C, Nord M, Didon L, Eklof AC, Frenckner B. Gene expression analysis after prenatal tracheal ligation in fetal rat as a model of stimulated lung growth. J Pediatr Surg. 2009;44:720–728. doi: 10.1016/j.jpedsurg.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Murakami S, Nagaya N, Itoh T, Iwase T, Fujisato T, Nishioka K, Hamada K, Kangawa K, Kimura H. Adrenomedullin regenerates alveoli and vasculature in elastase-induced pulmonary emphysema in mice. Am J Respir Crit Care Med. 2005;172:581–589. doi: 10.1164/rccm.200409-1280OC. [DOI] [PubMed] [Google Scholar]
- 36.Nagaya N, Mori H, Murakami S, Kangawa K, Kitamura S. Adrenomedullin: angiogenesis and gene therapy. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1432–1437. doi: 10.1152/ajpregu.00662.2004. [DOI] [PubMed] [Google Scholar]
- 37.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 38.Ogata Y. Bone sialoprotein and its transcriptional regulatory mechanism. J Periodontal Res. 2008;43:127–135. doi: 10.1111/j.1600-0765.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 39.Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan GQ, Ren HZ, Zhang SF, Wang XM, Wen JF. Expression of semaphorin 5A and its receptor plexin B3 contributes to invasion and metastasis of gastric carcinoma. World J Gastroenterol. 2009;15:2800–2804. doi: 10.3748/wjg.15.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell PP, Wang CC, Horinouchi H, Shepherd K, Jacobson M, Lipson M, Jones R. Differential expression of fibroblast growth factor receptors 1 to 4 and ligand genes in late fetal and early postnatal rat lung. Am J Respir Cell Mol Biol. 1998;19:563–572. doi: 10.1165/ajrcmb.19.4.2994. [DOI] [PubMed] [Google Scholar]
- 42.Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, Mariani TJ. Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. Am J Respir Crit Care Med. 2010;181:838–850. doi: 10.1164/rccm.200904-0544OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tschanz SA, Damke BM, Burri PH. Influence of postnatally administered glucocorticoids on rat lung growth. Biol Neonate. 1995;68:229–245. doi: 10.1159/000244241. [DOI] [PubMed] [Google Scholar]
- 44.Untergasser G, Gander R, Lilg C, Lepperdinger G, Plas E, Berger P. Profiling molecular targets of TGF-beta1 in prostate fibroblast-to-myofibroblast transdifferentiation. Mech Ageing Dev. 2005;126:59–69. doi: 10.1016/j.mad.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Usui H, Shibayama M, Ohbayashi N, Konishi M, Takada S, Itoh N. Fgf18 is required for embryonic lung alveolar development. Biochem Biophys Res Commun. 2004;322:887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- 46.Vadivel A, Abozaid S, van Haaften T, Sawicka M, Eaton F, Chen M, Thebaud B. Adrenomedullin promotes lung angiogenesis, alveolar development, and repair. Am J Respir Cell Mol Biol. 2009;43:152–160. doi: 10.1165/rcmb.2009-0004OC. [DOI] [PubMed] [Google Scholar]
- 47.Wagenaar GT, ter Horst SA, van Gastelen MA, Leijser LM, Mauad T, van der Velden PA, de Heer E, Hiemstra PS, Poorthuis BJ, Walther FJ. Gene expression profile and histopathology of experimental bronchopulmonary dysplasia induced by prolonged oxidative stress. Free Radic Biol Med. 2004;36:782–801. doi: 10.1016/j.freeradbiomed.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Weibel ER, Cruz-Orive LM. Morphometric methods. In: Crystal G, West JB, editors. The Lung: Scientific Foundations. 2. Philadelphia: Raven; 1997. pp. 333–344. [Google Scholar]
- 49.Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- 50.White KE, Ding Q, Moore BB, Peters-Golden M, Ware LB, Matthay MA, Olman MA. Prostaglandin E2 mediates IL-1beta-related fibroblast mitogenic effects in acute lung injury through differential utilization of prostanoid receptors. J Immunol. 2008;180:637–646. doi: 10.4049/jimmunol.180.1.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitsett JA, Clark JC, Picard L, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT. Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. J Biol Chem. 2002;277:22743–22749. doi: 10.1074/jbc.M202253200. [DOI] [PubMed] [Google Scholar]
- 52.Whitsett JA, Perl AK. Conditional control of gene expression in the respiratory epithelium: A cautionary note. Am J Respir Cell Mol Biol. 2006;34:519–520. doi: 10.1165/rcmb.F310. [DOI] [PubMed] [Google Scholar]
- 53.Williams SA, Schwarzbauer JE. A shared mechanism of adhesion modulation for tenascin-C and fibulin-1. Mol Biol Cell. 2009;20:1141–1149. doi: 10.1091/mbc.E08-06-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolff JC, Wilhelm J, Fink L, Seeger W, Voswinckel R. Comparative gene expression profiling of post-natal and post-pneumonectomy lung growth. Eur Respir J. 2010;35:655–666. doi: 10.1183/09031936.00059709. [DOI] [PubMed] [Google Scholar]
- 55.Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]


