Table 1.
Classification of helical protein interfaces.
| Types of PPIs | PDB code, helix sequence | Arrangement of hot spot residues on the target helix | ΔΔG (kcal/mol)1 | ΔSASA2 | Known Inhibitors | Protein names Function |
|---|---|---|---|---|---|---|
| Interfaces with Binding Clefts | ||||||
|
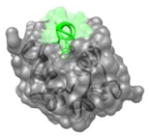
1YCR 19FSDLWKLLPE28 |
i | 3.8 | 131.1 | MDM2 (gray): coactivator p53 (green): transcriptional activator. Function:Apoptotic signaling, cell-cycle arrest |
|
| i + 4 | 4.4 | 136.7 | ||||
| i+ 7 | 2.0 | 73.4 | ||||
| (Avg) | (3.4) | (113.7) | ||||

|
1Y3A 5WYDFLM10 |
i | 4.2 | 168.3 | Giα 1 (gray): Guanine nucleotide-binding protein alpha-1 subunit KB752 (green): peptide Function: Signaling protein |
|
| i + 3 | 3.3 | 93.9 | ||||
| i +4 | 1.8 | 83.8 | ||||
| (Avg) | (3.1) | (115.3) | ||||
|
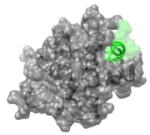
1BXL 578LAIIGDDI585 |
i | 2.7 | 102.2 | BCL-XL (gray): BAK peptide (green): Function: apoptosis |
|
| i + 3 | 2.2 | 87.3 | ||||
| i +7 | 1.2 | 79.3 | ||||
| (Avg) | (2.0) | (89.6) | ||||
|
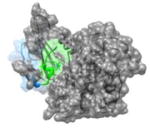
1JSU 85PEFYYR90 |
i | 2.9 | 122.1 | CDK2(gray): cyclin dependent kinase-2 P27(KIP1): kinase inhibitor (blue, helix in green) Function: Enzyme Kinase |
|
| i+1 | 4.9 | 116.8 | ||||
| i+3 | 1.1 | 64.6 | ||||
| (Avg) | (3.0) | (101.2) | ||||
| Extended Helical Interfaces | ||||||
|
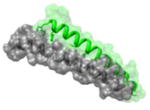
1AIK 628WMEWDREINNYTSLI642 |
i | 2.8 | 160.69 | HR1 (gray): N-terminal heptad repeat of HIV-1 GP41 HR2 (green): C-terminal heptad repeat of HIV-1 GP-41 Function: glycoprotein |
|
| i + 3 | 1.92 | 124.62 | ||||
| i+ 10 | 2.01 | 77.32 | ||||
| i+ 14 | 1.76 | 92.59 | ||||
| (Avg) | (2.12) | (113.8) | ||||
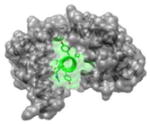
|
3BXK 1961IYAAMMIMEYYRQS1974 |
i | 1.9 | 130.49 | No known inhibitors | Ca2+-CaM (gray): calmodulin Cav 2.1 IQ (green): voltage gated calcium channels-Isoleucine-glutamine Function: Signaling protein |
| i + 1 | 1.6 | 105.38 | ||||
| i + 4 | 1.0 | 104.8 | ||||
| i + 6 | 3.1 | 111.97 | ||||
| i+ 10 | 2.7 | 103.28 | ||||
| i+11 | 2.0 | 107.88 | ||||
| (Avg) | (2.05) | (110.6) | ||||
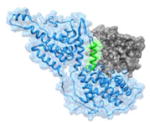
|
1NVW 930FGIYLTNILKTEEGN944 |
i | 1.64 | 43.73 | Ras (gray): guanine nucleotide binding protein Sos (blue, helix in green): Ras-specific guanine nucleotide exchange factor Function: signaling protein |
|
| i+6 | 1.11 | 57.75 | ||||
| i+13 | 1.10 | 57.98 | ||||
| i + 15 | 2.35 | 67.70 | ||||
| (Avg) | (1.55) | (56.79) | ||||
w
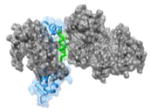
|
1K90 82EEEIREAFRVFDK94 |
i | 0.71 | 78.71 | No known inhibitors | EF(gray): Edema factor Calmodulin (blue, helix in green): Function: Anthrax Toxin |
| i+3 | 1.99 | 90.67 | ||||
| i+6 | 1.84 | 105.7 | ||||
| i+7 | 1.26 | 81.14 | ||||
| i+10 | 1.53 | 64.99 | ||||
| (Avg) | (1.47) | (84.24) | ||||
