Abstract
We describe the complete genome sequences of four closely related Hydrogenobaculum sp. isolates (≥99.7% 16S rRNA gene identity) that were isolated from the outflow channel of Dragon Spring (DS), Norris Geyser Basin, in Yellowstone National Park (YNP), WY. The genomes range in size from 1,552,607 to 1,552,931 bp, contain 1,667 to 1,676 predicted genes, and are highly syntenic. There are subtle differences among the DS isolates, which as a group are different from Hydrogenobaculum sp. strain Y04AAS1 that was previously isolated from a geographically distinct YNP geothermal feature. Genes unique to the DS genomes encode arsenite [As(III)] oxidation, NADH-ubiquinone-plastoquinone (complex I), NADH-ubiquinone oxidoreductase chain, a DNA photolyase, and elements of a type II secretion system. Functions unique to strain Y04AAS1 include thiosulfate metabolism, nitrate respiration, and mercury resistance determinants. DS genomes contain seven CRISPR loci that are almost identical but are different from the single CRISPR locus in strain Y04AAS1. Other differences between the DS and Y04AAS1 genomes include average nucleotide identity (94.764%) and percentage conserved DNA (80.552%). Approximately half of the genes unique to Y04AAS1 are predicted to have been acquired via horizontal gene transfer. Fragment recruitment analysis and marker gene searches demonstrated that the DS metagenome was more similar to the DS genomes than to the Y04AAS1 genome, but that the DS community is likely comprised of a continuum of Hydrogenobaculum genotypes that span from the DS genomes described here to an Y04AAS1-like organism, which appears to represent a distinct ecotype relative to the DS genomes characterized.
INTRODUCTION
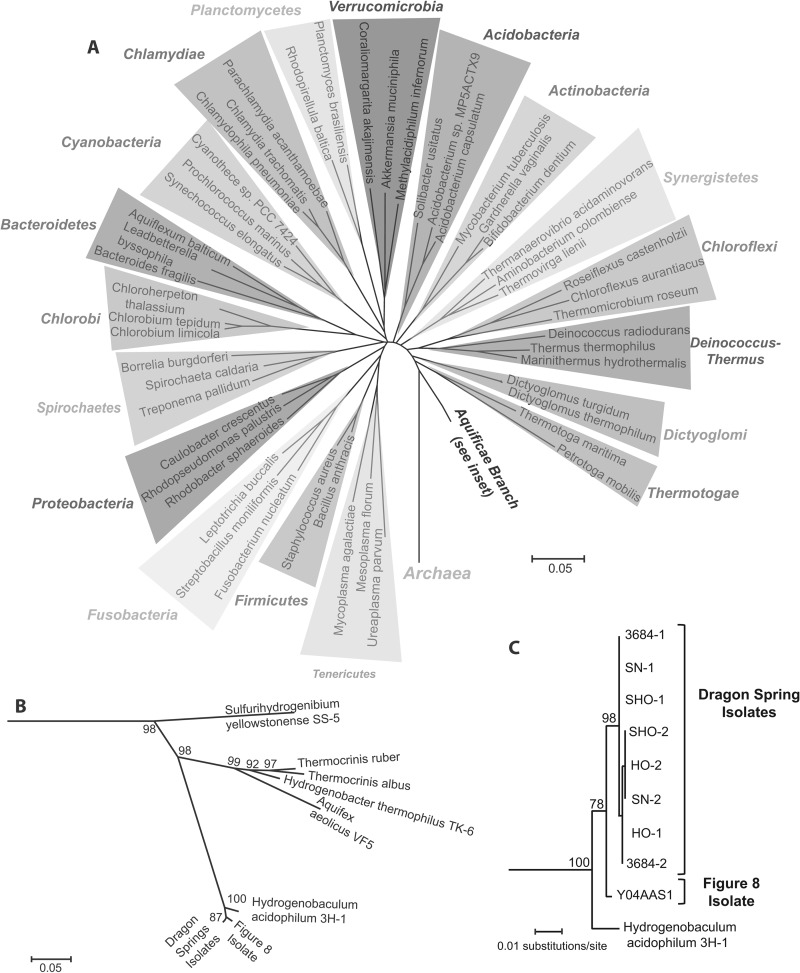
The genus species designation Hydrogenobaculum acidophilum was proposed by Stohr et al. (1) as a reclassification of Hydrogenobacter acidophilus due to the former's distant phylogenetic relationship to Hydrogenobacter, as well as its preference for very low pH. H. acidophilus was isolated from a solfataric mud in Japan and formally described as a Gram-negative rod using oxygen as an electron acceptor, hydrogen and reduced sulfur compounds as electron donors, and CO2 as the sole carbon source (1). H. acidophilum has an optimum temperature of 65°C and inhabits terrestrial geothermal environments. Phylogenetically, Hydrogenobaculum is a member of the Aquificae, a deeply branching phylum in the Bacteria domain (Fig. 1A).
Fig 1.
Phylogenetic position of Hydrogenobaculum strains examined in this study. (A) Neighbor-joining tree of representative bacterial phyla, with Aquificae shown near the base of the Bacteria domain. Note that some phylogeny projects place Thermotogae nearest to the base of the Bacteria (72, 73). (B) Magnified view of the Aquificales branch of the tree. (C) The Hydrogenobaculum branch within the Aquificales showing the positions of both 16S rRNA genes in each Hydrogenobaculum genome included in this study, except for strain Y04AAS1, wherein both 16S rRNA genes are identical. A complete list of the genes used for the main tree and the insets is given in Table S1 in the supplemental material, along with the IMG Gene Object identification numbers.
Thus far, most Hydrogenobaculum ecology and characterization work has been conducted in the Yellowstone National Park (YNP) geothermal complex (2–12), although this organism has also been found in Uzon Caldera, Kamchatka (13). In such environments, it has been inferred to occur based on molecular surveys in, or has been isolated from, sites with a temperature range of 50 to 91°C and a pH range of 1.02 to 5.75. Yellowstone Hydrogenobaculum ecology has been most extensively documented in the outflow channels of acid-sulfate springs (11) or acid-sulfate-chloride (ASC) springs (2–5, 9). YNP Hydrogenobaculum isolates have been shown to grow on H2, H2S, or thiosulfate as energy sources and CO2 as the sole carbon source (4–7, 12). In the outflow channels, geothermal source waters may contain significant concentrations of H2 and H2S and are supersaturated with CO2 relative to the atmosphere (4, 10, 11). Arsenite [As(III)] is also present at concentrations and flux levels that could easily support chemolithoautotrophic growth (10); however, Hydrogenobaculum isolates have thus far not been found to be capable of using As(III) as an energy source (4, 5). This bacterium will rapidly oxidize As(III) to arsenate but only in the absence of H2S (4, 5).
One ASC spring that has been extensively studied is Dragon Spring (DS), located in Norris Geyser Basin, YNP, WY (see Fig. S1A in the supplemental material). Its source waters exhibit a pH of ∼3.1 and contain millimolar levels of sulfate, chloride, and CO2 (measured as dissolved inorganic carbon) (10). A dominant feature of the DS outflow channel is a yellow S0 deposition zone that is home to a microbial community which rapidly consumes H2 and H2S (4) and fixes CO2 (2). Phylogenetic analysis of 16S rRNA genes PCR amplified and cloned from the microbial mat in this zone suggests that Hydrogenobaculum is a dominant member of this community (4, 9) and is consistent with later studies showing that Hydrogenobaculum isolates derived from this mat will grow on H2 or H2S singly or on both energy sources simultaneously (4).
Herein, we describe the complete genomes of four Hydrogenobaculum isolates obtained from DS that are nearly identical phylogenetically. We also compare these organisms to Hydrogenobaculum sp. strain Y04AAS1, which was isolated by the Reysenbach group (14) from an ephemeral stream connecting YNP geothermal features MVNN002 (referred to here as Figure 8 pool and abbreviated as F8) and Obsidian Pool-Prime (OP-P) in the Obsidian Pool Geothermal Complex, which is located in the YNP Mud Volcano area (see Fig. S1 in the supplemental material). These isolates are compared to each other and to available metagenomes from DS and OP-P. As revealed by their 16S rRNA genes and complete genome sequences, the DS and Y04AAS1 Hydrogenobaculum are phylogenetically related but nevertheless differ. We suggest that they represent different ecotypes. The data provide evidence of Hydrogenobaculum differentiation within the YNP geothermal complex, with part of this differentiation likely resulting from horizontal gene transfer.
Fig 8.
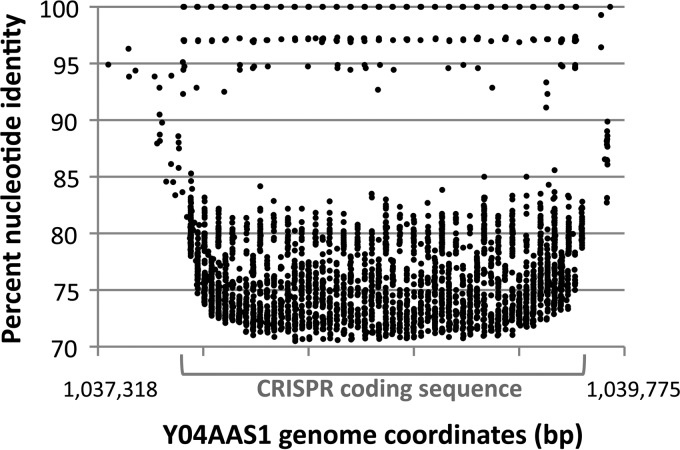
Fragment recruitment of the DS metagenome to the Y04AAS1 CRISPR coding region. The graph depicts each fragment within the DS metagenome that bears ≥70% identity (y axis) to the region of the Y04AAS1 genome that contains the CRISPR sequence (coordinates 1037710 to 1039665) and flanking DNA (x axis). DS DNA sharing complete identity with the Y04AAS1 CRISPR repeats is shown on the 100% identity line at the top. x axis tic marks denote 500 bp in length.
MATERIALS AND METHODS
Sample collection, culture isolation, and geochemical analysis.
DS (44°43′54.8″N, 110°42′39.9″W) is spring number NHSP106 in the Yellowstone National Park Thermal Inventory (http://www.rcn.montana.edu/resources/features/features.aspx?nav=11) and located in Norris Geyser Basin, YNP. Obsidian Pool (44°36′36.214″N, 110°26′19.758″W, entry MV007) and F8 (44°36′36.1″N, 110°26′21.8″W, entry MV006 and listed under feature name MVNN002) are components of the greater Obsidian Pool Geothermal Complex (see Fig. S1 in the supplemental material), which is located in the Mud Volcano area approximately 25.4 km from Norris Geyser Basin. OP-P is not a formal name in the Thermal Inventory Database but has been characterized and named by Spear et al. (15, 16). Aqueous geochemical analyses of DS have been reported previously (4, 10, 17), and sampling methods and protocols to analyze the aqueous constituents of F8 waters were described by Shock et al. (18). Samples 990723K and 000628B from that study were collected at F8, as was sample 040709B, reported in reference 19.
Hydrogenobaculum pure cultures were isolated as described previously (4). Genomic DNA was extracted from these cultures as described previously (4) and then amplified by multiple displacement amplification (MDA) using protocols described in detail in reference 20, including UV treatment of MDA reagents to eliminate amplification of contaminating DNA (21). The genome sequence of Hydrogenobaculum sp. strain Y04AAS1 was reported in reference 14. This organism was isolated from an ephemeral stream that derives from an outflow channel that drains the F8 pool. Depending on seasonal conditions, this ephemeral stream may form an aqueous bridge that connects F8 and OP-P (see Fig. S1 in the supplemental material). At the time of Y04AAS1 isolation, pool F8 was the primary source of nutrients and dissolved chemicals important to the Y04AAS1 environment, so we consider Y04AAS1 to be a member of the F8 microbial community.
Metagenome sequence preparation.
Metagenome analysis involved two of the 20 different YNP geothermal features that were part of the Yellowstone Metagenome Project (http://www.jgi.doe.gov/sequencing/why/99208.html). Protocols for DNA extraction, library construction, random shotgun sequencing, and sequence assembly were previously published (22). Metagenome sequences for the DS and OP-P sites can be found in the Integrated Microbial Genomes with Microbiomes Database (http://img.jgi.doe.gov/cgi-bin/m/main.cgi). The Taxon Object identifiers are 2014031004 and 2016842005, respectively.
Sequencing strategy.
Draft genomes of the Hydrogenobaculum DS genomes were generated at the DOE Joint Genome Institute (JGI) using a combination of Illumina GAii (23) and 454 Titanium (24) technologies, while manual finishing was performed at Los Alamos National Laboratory. The 454 Titanium standard data and the 454 paired-end data were assembled with Newbler version 2.3. The Newbler consensus sequences were computationally shredded into 2-kb overlapping fake reads (shreds). Illumina sequencing data were assembled with VELVET, version 0.7.63 (25), and the consensus sequences were computationally shredded into 1.5-kb overlapping shreds. We integrated the 454 Newbler consensus shreds, the read pairs in the 454 paired-end library, and the Illumina VELVET consensus shreds using parallel phrap, version SPS-4.24 (High Performance Software, LLC). The software Consed (26–28) was used in the subsequent finishing process. Illumina data were used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI (A. Lapidus, unpublished data). Possible misassemblies were corrected using gapResolution (C. Han, unpublished data), Dupfinisher (29), or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR, and by Bubble PCR (J.-F. Cheng, unpublished data) primer walks. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/.
Genomic analysis.
Genome analysis was carried out within the Integrated Microbial Genomes Database Expert Review (IMG-ER) system (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) (30). The MAUVE software program (31) was also used to search for single nucleotide polymorphisms between Hydrogenobaculum strains. Synteny alignments were made using the Artemis Comparison Tool (32), and CRISPR arrays were detected using the CRISPR recognition tool and CRISPRFinder (33, 34). Genomes were compared to metagenomes using fragment recruitment analysis (35). Average nucleotide identity (ANI) was calculated using the method described previously (36, 37). Percentage conserved DNA (PCD) was determined according to methods described previously (38).
Phylogenetic tree construction. (i) 16S rRNA genes.
16S rRNA genes were selected from Hydrogenobaculum and several other bacterial phyla (see Table S1 in the supplemental material for a complete list) and are available in the JGI Integrated Microbial Genomes Database. Sequences were aligned in CLUSTALW. The tree was generated using the neighbor-joining algorithm in MEGA (39).
(ii) Potassium-transporting ATPase kdpA.
The kdpA phylogenetic tree was constructed from gene sequences (see Table S2 in the supplemental material) that were obtained from the JGI Integrated Microbial Genomes Database (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) and selected from a variety of organisms representing various phyla. These sequences were aligned in MEGA (39) using the CLUSTALW algorithm and trimmed. The final tree was generated using the maximum likelihood algorithm in MEGA (39).
Nucleotide sequence accession numbers.
Gene annotations are available in GenBank: CP002726, Hydrogenobaculum sp. 3684; CP002809, Hydrogenobaculum sp. SHO; ADJO00000000, Hydrogenobaculum sp. SN; and CP004347, Hydrogenobaculum sp. HO.
RESULTS
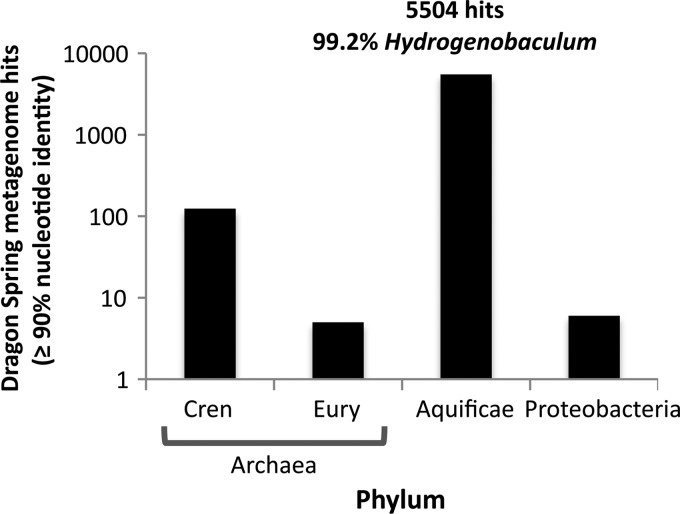
The DS hydrogenobacula described in this study were isolated from mat biomass taken approximately midway down the S0 deposition zone and in the middle of the outflow channel where water flow rates are maximal (see Fig. S1B in the supplemental material). BLAST analysis of the DS metagenome within the Integrated Microbial Genomes (IMG) Microbiome Database (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi) suggested that Hydrogenobaculum is a dominant component of the microbial community in this portion of the outflow channel. Based on the distribution of best UBLAST hits (R. C. Edgar, unpublished data) (≥90% identity) of 5,647 protein-coding genes, 5,504 genes are assigned to the Aquificae. Of the latter, 99.2% are annotated as being Hydrogenobaculum using the genome of Hydrogenobaculum strain Y04AAS1 as a reference (Fig. 2). At this same level of search stringency, other phyla/classes with very minor representation in this region of the DS outflow channel include the Crenarchaeota (124 hits), Euryarchaeota (5 hits), combined alpha-, beta-, and gammaproteobacteria (6 hits total), and Clostridia (2 hits).
Fig 2.
Prominent phyla in the Dragon Spring metagenome. Phylum/class representation is presented in log10 format to accommodate lower abundance organisms, and identification was based on ≥90% nucleotide identity to microbial reference genomes within the JGI-IMG database. Hydrogenobaculum identification within the Aquificae was also based on ≥90% nucleotide identity using the Hydrogenobaculum Y04AAS1 (GenBank accession no. CP001130.1) genome as a reference. At this level of search stringency, 79.06% of the DS metagenome sequences remain unassigned to any phylum.
Phylogenetic analysis of the 16S rRNA genes previously PCR amplified and cloned from DS found the Hydrogenobaculum to exhibit significant microdiversity (4, 9). The DS isolates examined herein are representative of this microdiversity in that they display very limited 16S rRNA gene sequence deviation (≥99.7% identity across 1,438 bp) (Fig. 1B and C). Each Hydrogenobaculum genome contains two 16S rRNA genes that within each DS isolate differ by three nucleotides in strain SHO, a single nucleotide in strain 3684, two differences in strain SN, and four changes between the two 16S genes in strain HO. In contrast, the two 16S rRNA genes in strain Y04AAS1 are identical. As a group, the DS strains branch separately from Y04AAS1 (Fig. 1C), differing by 10 to 15 nucleotides (99.30% to 98.96%). Only one 16S rRNA gene sequence is available for Hydrogenobaculum acidophilum isolated in Japan (1), and it places this organism basal to the two different Yellowstone phylotypes (Fig. 1C).
In terms of general genome features, each of the four closed DS genomes consists of a single circular chromosome with a length of 1,552,811 ± 155 bp (mean ± standard error, 0.01% variation across strains), and they are identical in G+C content (34.75%; Table 1). Of the ∼1,672 ± 4 predicted genes, 1,619 ± 4 are protein encoding, with the greatest variability noted for the pseudogene category (36 to 45 predicted in the four DS genomes; Table 1). Fig. S2 and Table S3 in the supplemental material list the distribution of genes into COG functional categories. The DS genomes are not identical, though the great majority of the differences are annotated as hypothetical proteins (see Table S4 in the supplemental material), and so it is not possible to determine if there are true functional differences between the DS isolates that could translate into separate ecotype designations. Analysis of single nucleotide polymorphisms (SNPs) among the DS genomes identified several functions that could potentially be affected by nonsynonymous changes in strains HO and SHO (see Table S5 in the supplemental material). Examples include (i) the incorporation of a translational stop signal in the GTP binding protein LepA, (ii) an S→L mutation in the heterodisulfide reductase subunit B, (iii) a G→V mutation in the hydrogenase ([NiFe] type) small-subunit HydA, and (iv) enhanced SNP activity in the flagellar biosynthesis/type III secretory pathway protein (see Table S5 in the supplemental material).
Table 1.
Summary of genome features for Dragon Spring strains 3684, HO, SHO, and SN and the F8 strain Y04AAS1
| Attribute | 3684 |
HO |
SHO |
SN |
Y04AAS1 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Value | % of total | Value | % of total | Value | % of total | Value | % of total | Value | % of total | |
| Size (bp) | 1,552,775 | 100 | 1,552,931 | 100 | 1,552,607 | 100 | 1,552,931 | 100 | 1,559,514 | 100 |
| G+C content (bp) | 539,616 | 34.75 | 539,676 | 34.75 | 539,566 | 34.75 | 539,673 | 34.75 | 543,444 | 34.85 |
| Coding region (bp) | 1,458,831 | 93.95 | 1,455,958 | 93.76 | 1,454,210 | 93.66 | 1,476,560 | 95.08 | 1,491,434 | 95.63 |
| Total no. of genes | 1,667 | 100 | 1,672 | 100 | 1,671 | 100 | 1,676 | 100 | 1,691 | 100 |
| No. of RNA genes | 53 | 3.18 | 53 | 3.17 | 53 | 3.17 | 52 | 3.1 | 51 | 3.02 |
| No. of rRNA operons | 6 | 0.36 | 6 | 0.36 | 6 | 0.36 | 6 | 0.36 | 6 | 0.35 |
| No. of protein-coding genes | 1,614 | 96.82 | 1,619 | 96.83 | 1,618 | 96.83 | 1,624 | 96.9 | 1,640 | 96.98 |
| No. of pseudogenes | 36 | 2.16 | 40 | 2.39 | 45 | 2.69 | 36 | 2.16 | 11 | 0.65 |
| No. of genes with function prediction | 1,322 | 79.3 | 1,324 | 79.19 | 1,206 | 72.17 | 1,329 | 79.3 | 1,194 | 70.61 |
| No. of genes in paralog clusters | 34 | 2.04 | 38 | 2.27 | 36 | 2.15 | 41 | 2.45 | 643 | 38.02 |
| No. of genes assigned to COGs | 1,337 | 80.2 | 1,337 | 79.96 | 1,336 | 79.95 | 1,338 | 79.83 | 1,262 | 74.63 |
| No. of genes assigned Pfam domains | 1,287 | 77.2 | 1,286 | 76.91 | 1,288 | 77.08 | 1,289 | 76.91 | 1,294 | 76.52 |
| No. of genes with signal peptides | 208 | 12.48 | 210 | 12.56 | 207 | 12.39 | 209 | 12.47 | 234 | 13.84 |
| No. of genes with transmembrane helices | 395 | 23.7 | 396 | 23.68 | 397 | 23.76 | 397 | 23.69 | 396 | 23.42 |
| CRISPR repeats | 7 | 7 | 7 | 7 | 1 | |||||
Comparing DS strains to Y04AAS1.
Several differences between the DS and Y04AAS1 genomes can be seen by comparing the general genome features (Table 1), COG functional categories (see Table S3 in the supplemental material), and specific annotated functions (see Tables S5 and S6 in the supplemental material). The DS genomes contain 78 annotated genes that are absent in Y04AAS1 (see Table S6). Hypothetical proteins or proteins of unknown function are again a prominent category (28 total), although there are several specific functions unique to the DS genomes. Potentially ecologically relevant genes include those coding for As(III) oxidation [As(III) oxidase large- and small-subunit genes, aioBA; see Table S6]. The absence of aioBA in the Y04AAS1 genome may correlate to the lower As(III) levels observed in the F8 geothermal feature, which also differs from DS with respect to SO4−2, H2S, and Fe2+ (Table 2). During the course of this study, DS chemistry was remarkably stable (3), whereas pH varied by nearly a full unit in the Y04AAS1 isolation site at two sampling times in the summer of 2004 (Table 2). We also note that pH ranged between approximately 2.6 and 5 during earlier sampling in the 1999 field season (results not shown). At the time of sampling of the DS biomass for DNA extraction, the source water temperature was identical to that of pool F8 (Table 2).
Table 2.
Comparable aqueous chemical features of Dragon Spring and F8 geothermal featuresa
| Site | Sampling date(s) | Temp (°C) | pH | Analyte |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Na+ (mM) | SO42− (mM) | Arsenic (μM) | H2S (μM) | Fe2+ (μM) | NO3− (μM) | ||||
| Dragon Spring | November 1999; July 2006 | 63 | 3.1 | 18.0 | 1.3 | 35 | 60 | 65 | 17.6–18.5b |
| Figure 8 pool | 9 July 2004 | 65 | 3.1 | 18.4 | 4.14 | 9.4 | 0.5 | 6.8 | BDc |
| Figure 8 pool | 20 July 2004 | 65 | 4.0 | 20.9 | 3.63 | 9.3 | 4.1 | 19 | BD |
Dates refer to when the geochemical analyses were conducted. Unless otherwise noted, Dragon Spring data are from Langner et al. (10), although they are in agreement with that observed in 2006 and reported by D'Imperio et al. (4).
Data from Ackerman (40), sampled in 2003 to 2005.
BD, below detection.
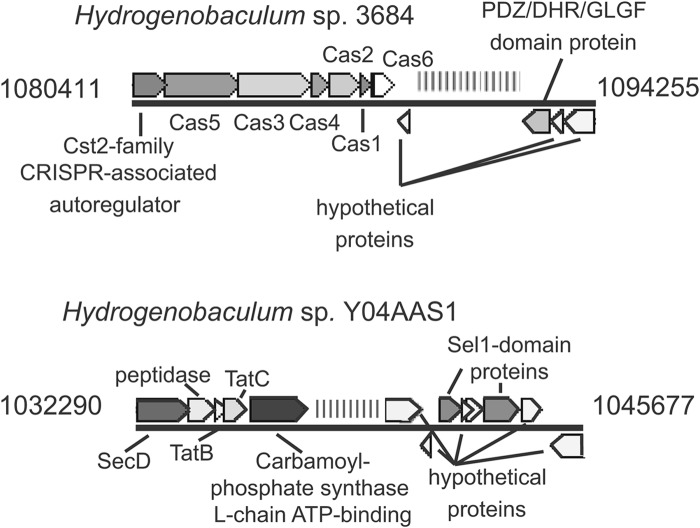
Another difference between the DS and F8 Hydrogenobaculum strains involves CRISPR regions (Fig. 3), loci that are viewed to derive from phage infection and that contribute to antivirus immunity (41–51). The DS strains each encode seven CRISPR regions, which are scattered throughout the genomes (Fig. 4). Within each DS strain, each CRISPR locus differs considerably, but each is nearly identical to its corresponding locus in the other DS strains. For example, CRISPR 1 and CRISPR 2 of Hydrogenobaculum SHO differ greatly, but they are very similar to CRISPR 1 and CRISPR 2, respectively, in Hydrogenobaculum SN, HO, and 3684. In contrast, the Y04AAS1 genome encodes a single CRISPR region with spacer sequences that bear no resemblance to any in the DS Hydrogenobaculum strains. In addition, Y04AAS1 and the DS strains differ in the presence of cas genes. There are at least two prominent categories of cas genes: “core” and “subtype” (42, 44, 46, 47, 50). Core cas genes found in the DS Hydrogenobaculum include Cas1 and Cas2, which are thought to be universal, and Cas3, Cas4, Cas5, and Cas6, which are not universal but still quite common (42, 44, 50). Subtype Cas proteins found in the DS Hydrogenobaculum include Cst2 (Fig. 3), which is associated with Thermotoga species (50). The sequences of the cas operons are nearly identical in all four DS strains, with the exception of strain SN, where an “A” nucleotide is omitted after position 341508 and again after position 1083075. The Y04AAS1 genome does not contain any annotated cas genes.
Fig 3.
Comparison of Hydrogenobaculum CRISPR loci in Dragon Spring strain 3684 and F8 strain Y04AAS1. CRISPR 7 from DS strain 3684 is similar to CRISPR 7 from all other DS isolates and is pictured above the one CRISPR locus from Hydrogenobaculum sp. Y04AAS1. CRISPR 7 was selected for comparison because it is the longest and most complex CRISPR region within the DS Hydrogenobaculum genomes. The array of spacers and repeats is denoted by vertical stripes, and labels indicate the annotation of flanking genes. Of particular note, strain 3684 encodes several “core” cas (CRISPR-associated) genes (44, 47, 50), whereas Y04AAS1 does not. CRISPR arrays were detected using the CRISPR recognition tool (33) and CRISPRFinder (34).
Fig 4.
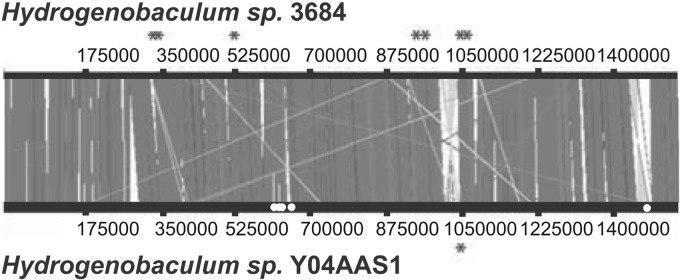
Synteny between the Dragon Spring and Y04AAS1 Hydrogenobaculum genomes. Dragon Spring genomes are represented by strain 3684. Asterisks denote the approximate location of the CRISPR loci in each genome. Dark gray denotes syntenic regions, pale gray highlights large inversions, and gaps (white spaces) illustrate regions that contain different DNA sequences. White dots indicate regions of the Y04AAS1 genome predicted to be acquired from Thermodesulfobium narugense via horizontal gene transfer.
The Y04AAS1 genome is roughly 6,700 bp longer than the DS genome and differs in virtually every COG category. While most differences are quantitatively small, there are some examples where the Y04AAS1 genome differs by ±20% in number of genes per COG category. Examples include (i) replication, recombination, and repair; (ii) cell cycle control; (iii) defense mechanisms; (iv) signal transduction mechanisms; and (v) intracellular trafficking and secretion (Table 1). More specifically, the Y04AAS1 genome is annotated to contain 84 genes that are absent in the DS isolates (see Table S6 in the supplemental material). Of these, 46 are either hypothetical proteins or proteins of unknown function. There are specific functions that appear unique to Y04AAS1, including nitrate and nitrite dissimilatory reduction (biosynthesis of the cytochrome d1 heme region and nirE required for nitrite reduction), nitric oxide reductase (norZ apoprotein), thiosulfate metabolism (soxB, soxY, soxZ), and two proteins annotated as having a function associated with heavy metal transport/detoxification. One of the latter genes (locus tag HY04AAS1_1211) shares 42% amino acid identity with the mercuric transport protein MerT from Hydrogenovirga sp. 128-5-R1-1, and the other (HY04AAS1_1212) shares a 64% amino acid identity with the mercuric reductase from Hydrogenobacter thermophilus TK-6, as determined by a BLAST search (Gene Object identifiers 642751020 and 642751021, respectively; see Table S5 in the supplemental material).
Synteny among the DS genomes is exceptionally strong. The average nucleotide identity (ANI) across all DS genomes ranges from 99.996% to 99.999%, averaging 99.998 ± 0.001% (mean ± standard deviation). In contrast, the DS genomes are noticeably different from the F8 isolate Y04AAS1. The ANI for comparisons between DS and Y04AAS1 is 94.764 ± 0.007%, and the PCD is 80.552 ± 0.019% (Table 3).
Table 3.
Average nucleotide identity and percentage conserved DNA comparisons for the Dragon Spring and F8 genomes
| Strain | Avg % nucleotide identitya |
||||
|---|---|---|---|---|---|
| DS genomes |
F8 genome, Y04AAS1 | ||||
| SN | HO | SHO | 3684 | ||
| SN | 99.998 (0.978) | 99.999 (0.978) | 99.998 (0.979) | 94.761 (0.889) | |
| HO | 99.997 (0.980) | 99.996 (0.980) | 94.761 (0.891) | ||
| SHO | 99.998 (0.983) | 94.761 (0.894) | |||
| 3684 | 94.774 (0.895) | ||||
| SN | 99.996 | 99.952 | 99.961 | 80.541 | |
| HO | 99.954 | 99.958 | 80.542 | ||
| SHO | 99.978 | 80.580 | |||
| 3684 | 80.546 | ||||
| 80.552 ± 0.019 | |||||
For each genome comparison, the average percent identity is followed parenthetically by the proportion of the smallest genome aligned. Percentage conserved DNA comparisons are also summarized among DS genomes and the F8 genome.
Comparison of DS and Y0AAS1 genomes to the DS metagenome.
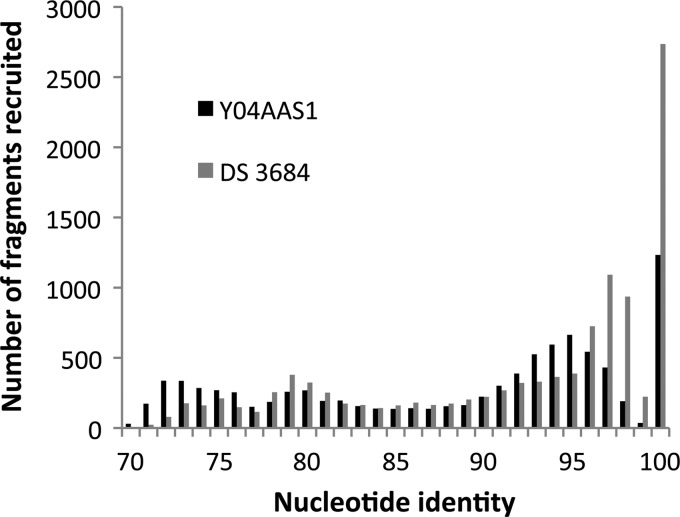
DS genomes (strain 3684 as representative) and the Y04AAS1 genome were used as references for a comparative fragment recruitment analysis of the DS metagenome (Fig. 5). Within the range of 70 to 100% nucleotide identity, the DS genome accumulates 11,127 hits compared to 9,091 hits for the Y04AAS1 genome. At ≥95% DNA identity, the DS strains recruit nearly twice as many reads from the DS metagenome as Y04AAS1. This suggests the DS genomes are more representative of the hydrogenobacula within Dragon Spring than Y04AAS1. Recruitment analysis based on Y04AAS1 KEGG functional categories found that energy, carbohydrate, and amino acid metabolisms were the dominant functions, sharing 100% identity with the DS metagenome (results not shown).
Fig 5.
Fragments of the Dragon Spring metagenome recruited by the DS strain 3684 and F8 strain Y04AAS1 genomes as references.
Evidence of horizontal gene transfer.
We used the Putatively Horizontally Transferred Genes tool within the Integrated Microbial Genomes Database (https://img.jgi.doe.gov/cgi-bin/er/main.cgi) to examine the basis for the ANI and PCD differences between the DS and Y04AAS1 genomes. Of the 84 genes that are unique to strain Y04AAS1 relative to the DS genomes (see Table S6 in the supplemental material), 43 are predicted to be acquired via horizontal gene transfer (HGT) (see Table S7 in the supplemental material). Of these, nearly half (20/43) are suggested to have come from an organism that is a thermophile, an acidophile, or an organism having both features (see Table S7). Of particular interest are genes predicted to have been acquired from Thermodesulfobium narugense (see Table S7), which has a G+C content of 34.58% and thus quite similar to that of Y04AAS1 (34.85%; Table 1). In total, of the 35,119 bp of putative HGT DNA, nearly half (48%, 16,965 bp) are predicted to come from T. narugense. Further, significant segments of the putative T. narugense DNA occur at a discrete location in the Y04AAS1 genome (see Fig. 4), consistent with an intermolecular recombination event. Y04AAS1 genes that share homology with T. narugense include hypothetical proteins, an entire K+-transporting ATPase operon, and three adjacent genes coding for glycosyl transferase activity (see Table S7).
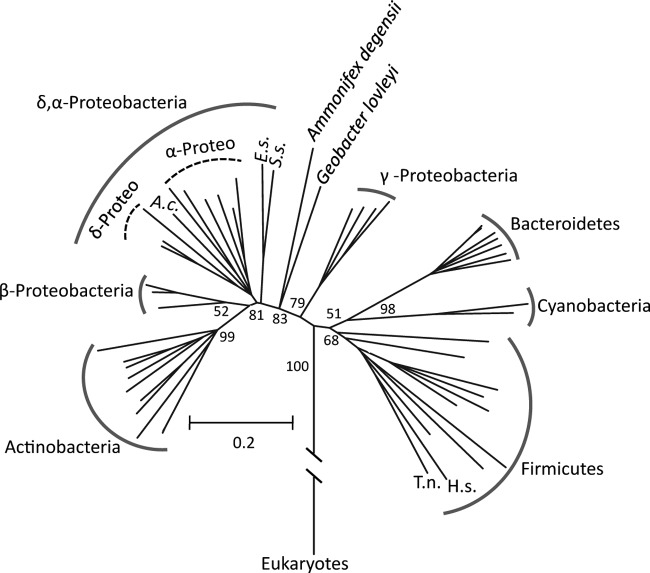
To further assess potential HGT, we examined the phylogenetic relatedness of the K+-transporting ATPase subunit A genes from Y04AAS1 and several other organisms. These genes belong to the kdpA superfamily of potassium-transporting ATPases and are referred to here as kdpA. A maximum likelihood analysis shows that the kdpA genes from a variety of organisms cluster into discrete clades that primarily correspond to the organism's 16S rRNA gene affiliation (Fig. 6; kdpB clade structure is essentially the same [data not shown]). The kdpA genes from Y04AAS1 and T. narugense are very closely related and fall within the Firmicutes (Fig. 6). T. narugense is currently described as a firmicute by Mori and Hanada (52), although its exact taxonomic position remains unsettled (S. Hanada, personal communication). The original description of T. narugense placed this organism most closely to the candidate phylum OP9 (53), first proposed based on environmental clones PCR amplified from Obsidian Pool, which is connected to OP-P (see Fig. S1 in the supplemental material). Regardless of whether T. narugense is a firmicute or a member of the candidate phylum OP9, phylogenetic placement of the Hydrogenobaculum Y04AAS1 kdp genes departs considerably from the phylum Aquificae in which these particular kdp genes appear to be lacking and thus is consistent with these genes being acquired horizontally.
Fig 6.
Evidence that Y04AAS1 acquired the potassium-transporting ATPase subunit A gene (kdpA) via horizontal gene transfer from Thermodesulfobium narugense. In some cases, full species names were abbreviated to conserve space. The kdpA alleles primarily grouped according to 16S rRNA gene phyla, though there were a few exceptions: Eggerthella sp. (E.s., phylum Actinobacteria) and Synergistes sp. (S.s., phylum Synergistetes), Acidobacterium capsulatum (A.c., phylum Acidobacteria), Ammonifex degensii (phylum Firmicutes), and Geobacter lovleyi (phylum Deltaproteobacteria). Additional abbreviations include T.n., Thermodesulfobium narugense; and H.s., Hydrogenobaculum sp. The genes used to construct this tree are listed in Table S8 in the supplemental material.
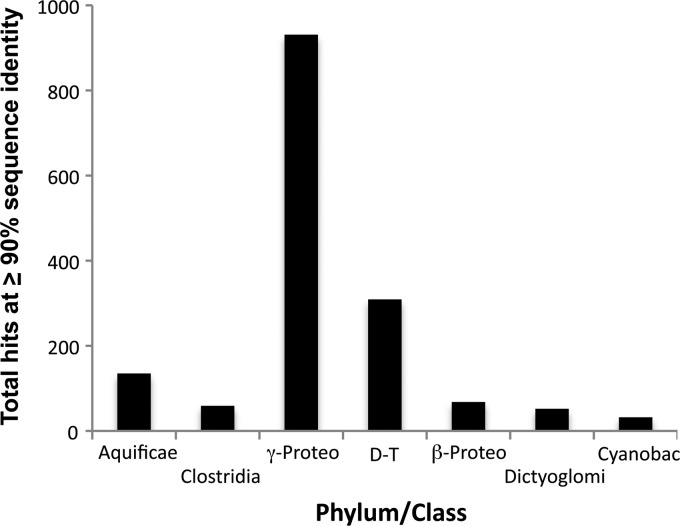
We next sought to establish evidence of T. narugense occurring in OP-P or Obsidian Pool, so as to place the potential HGT donor organism within this geothermal pool complex. T. narugense is an H2-oxidizing, sulfate-respiring, anaerobic bacterium having an optimal pH of 5.5 to 6.0 and an optimal temperature of 55°C (53). These critical physiologic features are similar to the environmental conditions reported for OP-P (pH 5.7, 325 nM H2, 2.4 mM SO42−) and Obsidian Pool (pH 6.5, 133 nM H2, 1.55 mM SO42−) (16). Analysis of the OP-P metagenome (OPP_17 in the YNP Metagenome, http://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2016842005) revealed evidence of T. narugense-like organism(s). Using tools available within IMG/MER (http://img.jgi.doe.gov/cgi-bin/mer/main.cgi), a fragment recruitment analysis was performed at high stringency (≥90% nucleotide identity) using all genomes within the IMG/MER as references. This analysis suggested that the Gammaproteobacteria and Thermus are significant taxa in OP-P; however, both the phylum Aquificae and the class Clostridia (phylum Firmicutes) are also predicted to be represented (Fig. 7). Hydrogenobaculum homologous genes represent 21% of the Aquificae-annotated genes, and Thermodesulfobium accounts for 90% of the Clostridia homologous DNA. These totals represent 1.8% and 3.4%, respectively, of the OP-P metagenome hits represented in Fig. 7.
Fig 7.
Identification of prominent phyla in the Obsidian Pool Prime metagenome (D-T, Deinococcus-Thermus). Identification was based on ≥90% nucleotide identity to microbial reference genomes within the JGI-IMG database. At this level of search stringency, 95.7% of the Obsidian Pool Prime metagenome sequences remained unassigned to any phylum.
Finally, to further explore the basis for the similarity between Y04AAS1 and the DS Hydrogenobaculum populations (Fig. 5) and to ascertain whether Y04AAS1 is present in DS, the DS metagenome was searched for Y04AAS1-specific marker genes. The genes selected were those unique to Y04ASS1, such as the putative HGT genes (see Tables S6 and S7 in the supplemental material) and the CRISPR locus (Fig. 3). A BLAST search using the putative HGT-derived T. narugense-like genes in separate queries identified a total of 30 hits to eight of the 10 genes, with low-to-complete coverage and with high identity and quality scores (Table 4). Recruitment of the DS metagenome to the Y04ASS1 CRISPR (Fig. 8) identified one complete set of repeats identical to the Y04ASS1 CRISPR, another that shares 97% identity, and then a very large number of repeats that are of significantly lower identity (Fig. 8). The spacers were diverse and exhibited substantially poorer matches (predominantly <85%) (Fig. 8).
Table 4.
BLAST search analysis of DS metagenome for Y04AAS1 marker genes believed to be derived from Thermodesulfobium narugense
| Y04AAS1 gene identifiera | Annotation | Query length (bp) | No. of hits | Query coverage (%) | Avg identity (%) | E value range |
|---|---|---|---|---|---|---|
| 642750486 | Hypothetical protein | 183 | 0 | NA | NA | NA |
| 642750487 | Hypothetical protein | 1,755 | 2 | 63 | 96 | 0 to 2.0e−152 |
| 642750498 | Diguanylate cyclase/phosphodiesterase | 1,902 | 3 | 100 | 96 | 0 to 1.0e−45 |
| 642750516 | K+-transporting ATPase, A subunit | 1,731 | 5 | 66 | 93 | 9.0e−102 to 4.0e−56 |
| 642750517 | K+-transporting ATPase, B subunit | 2,055 | 1 | 38 | 95 | 0 |
| 642750518 | K+-transporting ATPase, C subunit | 597 | 0 | NA | NA | NA |
| 642750519 | Osmosensitive K+ channel His kinase sensor | 1,932 | 3 | 20 | 97 | 0 to 2.0e−29 |
| 642751394 | Methyltransferase type 12 | 2,241 | 4 | 62 | 99 | 0 to 2.0e−10 |
| 642751395 | Glycosyl transferase family 2 | 2,241 | 4 | 98 | 95 | 0 to 6.0e−12 |
| 642751396 | Glycosyl transferase family 2 | 948 | 3 | 98 | 96 | 0 to 6.0e−12 |
The Y04AAS1 gene identifier refers to the IMG database.
DISCUSSION
Acidic hot springs are a major type of geothermal feature in YNP (54), the world's largest, most complex, yet best-characterized geothermal complex. ASC springs are well represented, particularly in Norris Geyser Basin. Within the outflow channels of the ASC springs such as DS, the microbial community is relatively simple (Fig. 2). The heavy representation of the Aquificae in the DS metagenome did not result from bias due to unbalanced reference genome representation. The Aquificae are represented by only 13 genomes, whereas hundreds of genomes comprise the other phyla used as references to analyze DS (BLAST criterion of ≥90% nucleotide identity). Exceptionally strong Hydrogenobaculum representation in the metagenome is completely consistent with our previous 16S rRNA gene cloning at this site, which demonstrated that within the domain Bacteria, Hydrogenobaculum was overwhelmingly dominant (4, 9). Consequently, it is evident that Hydrogenobaculum constitutes a significant component of the microbial community in ASC geothermal features and thus warrants continued examination to determine its role in nutrient and elemental cycling.
As defined by the 16S rRNA gene, Hydrogenobaculum microdiversity in YNP is substantial (4, 9), though the ecological or evolutionary significance of such microdiversity is not understood for this bacterium or others for which significant 16S rRNA gene microdiversity has been reported (e.g., the SAR-11 Pelagibacter complex [55]). The genome of Hydrogenobaculum sp. strain Y04AAS1 contains two identical 16S rRNA genes, implying that each novel sequence encountered in a PCR-generated environmental clone library accounts for a separate population. However, the current study illustrates that potentially as much as half of the Hydrogenobaculum 16S rRNA gene microdiversity observed in YNP derives from variability between rRNA operons within the same organism (Fig. 1C). The distinct clustering of the DS 16S rRNA genes (Fig. 1C) corresponds to highly similar genomic composition, suggesting that limited functional differences may be inferred from 16S rRNA gene clone libraries that display similarly close relatedness. The intragenomic microdiversity notwithstanding, an ecological explanation probably exists for the very significant microdiversity of the Hydrogenobaculum 16S rRNA genes observed in environmental clone libraries. More investigations are needed to determine how and why this microdiversity occurs and is maintained.
While highly similar, the different DS genomes nevertheless are predicted to contain unique genes relative to each other (see Table S4 in the supplemental material). Most of these strain-specific genes are annotated as coding for hypothetical proteins, which is a trend also observed when closely related Escherichia coli strains are compared to each other (56). This makes it difficult to predict specialized function among these isolates that might correlate with ecological behavior or fitness (i.e., ecotype designation). Hypothetical proteins are also among the genes affected by SNPs in the DS genomes. Other differences include silent mutations in transposases and nonsynonymous changes in an [NiFe] hydrogenase small subunit (HydA) and heterodisulfide reductase. The change in the latter occurs in a poorly conserved region of the gene and, as such, might be inconsequential. The nonsynonymous change in HydA occurs in a well-conserved region, as determined based on the amino acid sequence and crystal structure of the Allochromatium vinosum functional [NiFe] hydrogenase (UniProtKB D3RV29). Thus, while the HydA mutation might influence function, it was found in DS strains HO and SHO, both of which were found to grow on H2 as a sole energy source (4). The HydA mutation could influence enzyme kinetics (e.g., increased Km), potentially affecting the relative competitiveness of HO and SHO in regions of the outflow channel where H2 occurs at low concentrations due to off-gassing (4). In previous work, we reported that the DS strains exhibit different ecologically relevant phenotypes (4); i.e., strains 3684 and SHO could grow on either H2 or H2S, strain SN could grow on H2S but not H2, and strain HO grew on H2 but not H2S. A genotypic basis for these differences was not readily apparent when assessing SNPs in genes annotated for relevant functions (hydrogenase or sulfide quinone reductase), though there were several differences involving hypothetical proteins in SN relative to the other strains (see Table S3 and S4 in the supplemental material). To conclude the absence of, or mutations in, hypothetical proteins as the basis for failure of SN to grow on H2 implies that these hypothetical proteins play a role(s) in H2 metabolism that has not yet been identified.
While the DS isolates are genetically extremely similar, they are measurably different from the F8 strain and the type strain H. acidophilum. Biogeographical patterns correlate with 16S rRNA gene phylogeny (57, 58), and so it is not necessarily surprising to observe the phylogenetic separation of the YNP isolates from the Japanese isolate H. acidophilum (Fig. 1C). However, the DS strains form a cluster that separates from the F8 strain with strong bootstrap support (Fig. 1C), and this same topology pattern is observed when we use 40 single-copy protein families to construct a tree based on more comprehensive information (aligned using the Pfam Hidden Markov Models; results not shown). The potential taxonomic significance of this phylogenetic branching pattern was investigated using ANI and PCD. These are quantifiable genome metrics proposed to replace 70% DNA-DNA hybridization (37, 38), which has been the accepted standard for determining species-level taxa (59). The ANI for comparisons of DS and F8 of 94.76% (Table 3) is squarely at the ANI threshold (ANI ≤ 95%) that correlates with 70% DNA-DNA hybridization to distinguish a species (37, 38). The PCD value of ∼80%, derived from comparing the DS and F8 genomes, also demonstrates YNP organismal differences, but it does not meet the requisite threshold for species designation (<69%) (38). Thus, from analyses comparing the 16S rRNA gene and genome-wide properties of the DS and F8 strains, it is evident that they differ, though perhaps not sufficiently so as to unambiguously recommend a species designation based on current views and definitions.
Alternatively, the differences between these related bacteria might reflect an ecotype relationship as defined by Cohan (60), wherein organisms cluster according to their genetic, phenotypic, and ecological characteristics. These criteria appear satisfied by the Hydrogenobaculum isolates discussed here. The strains cluster phylogenetically (Fig. 1C), and this clustering correlates with physiological and phenotypic differences (Table 1; see also Tables S3 to S7 in the supplemental material). For example, Y04AAS1 possesses the genetic capacity to respire nitrate and can use thiosulfate as an energy source and detoxify Hg (see Table S6) (61). Conversely, the DS hydrogenobacula lack these determinants but can oxidize As(III) (see Table S6) (5, 17). As such, phenotypic differences also distinguish the DS isolates from the F8 isolate.
The ecology of DS and F8 geothermal pools differs (Table 2), but it becomes somewhat difficult to directly link these different ecological settings with phenotype and/or genotype. Comparable chemical data for these geothermal features (Table 2) is limited for the specific constituents that now, subsequent to organism isolation, are of interest as a result of these genomic analyses and comparisons. Based on multiple studies conducted before, during, and after the isolation of the DS isolates (3, 4, 10, 17), the DS geochemistry appears extremely stable. However, in 2004, the same year that strain Y04AAS1 was isolated (A.-L. Reysenbach, personal communication), the F8 pool geochemistry was variable, with major selective features such as pH and H2S changing nearly an order of magnitude within the same summer sampling season (Table 2). Furthermore, NO3− and NO2− levels were below detection in F8, and so F8 chemistry cannot be linked to the ability of Y04AAS1 to respire nitrate as suggested by its genotype. As(III) oxidation and the in situ expression of aioA (previously referred to as aoxB) in Hydrogenobaculum are well-documented features of the DS outflow channel microbial mats (3, 10, 17), and so it is not surprising to find the As(III) oxidase structural genes (aioBA) in the DS genomes. Arsenic concentrations in F8 were about 3-fold lower than at DS (Table 2) and potentially could correlate with the apparent lack of As(III) oxidase genes (aioBA or arxA) in the F8 strain. However, it is difficult to firmly correlate As concentrations and the presence/absence of As(III) oxidase genes, because Inskeep et al. (62) and Hamamura et al. (63) detected aioA (referred to as aroA) in various environments containing a range of As concentrations, including those that contained far less As than observed in F8. The apparent lack of Hg resistance genes in the DS genomes may be inconsequential, as Boyd et al. (2) documented very low mercury levels in DS waters (HgTotal = 38 ng liter−1 and MeHg+ below a detection limit of 0.025 ng liter−1).
Additional differences between the DS and F8 strains were observed in the CRISPR regions. CRISPR loci are stretches of semirepetitive DNA sequences that result when a virus, plasmid, or other source of foreign DNA infects a prokaryote (36, 41, 42, 47, 49, 50). They are helpful for characterizing the viral community in a host organism's habitat (51, 64) and also provide a genetic marker with which to distinguish individual strains within a larger community (51). The complete lack of CRISPR homology between the DS and F8 genomes indicates the presence of similar, but still different, virus populations in the respective YNP geothermal features from which these strains were collected. This result was not unexpected, however, given the diversity of viruses discovered thus far in YNP (65–67) and the complete lack of Hydrogenobaculum virus information.
The DS metagenome analysis illustrated that while both DS and F8 genomes can efficiently recruit from this metagenome, the DS genomes appear to better represent the Hydrogenobaculum found at that same location (Fig. 5). Hydrogenobaculum strains other than those represented by the DS genomes occur in DS, but three current lines of evidence suggest that strain Y0AAS1, per se, may not actually be present in DS. First, the 16S rRNA gene sequence for Y04AAS1 demarcates this strain as phylogenetically separate from the DS strains (Fig. 1C) but has yet to be found in any DS 16S rRNA gene PCR clone libraries (4, 9), including 333 near-full-length Hydrogenobaculum 16S rRNA gene clones generated as part of the YNP metagenome analysis of DS (203 of which were unique). Second, when the T. narugense-like genes viewed to have been acquired by Y04AAS1 via HGT were searched against the DS metagenome, 30 BLAST hits specifically matched eight of the 10 genes exhibiting significant matches, though still deviating significantly from Y04AAS1 (Table 4). And third, BLAST searches of the DS metagenome using the Y0AAS1 CRISPR region (2,458 bp) as the query failed to identify the lone Y04AAS1 CRISPR as defined by its spacer sequences. Rather, we detected a Y04AAS1-like CRISPR that shares identical repeat sequences but lacks the same spacer sequences and thus infers exposure to a different population(s) of phage relative to those represented by the spacers in the DS strains (Fig. 8). The significant spacer heterogeneity (Fig. 8) suggests that the Hydrogenobaculum phages in DS are rapidly diversifying and that spacer sequence types may serve as Hydrogenobaculum population indicators in a fashion similar to that observed in Leptospirillum biofilms (51). As a group, the DS spacers appear to cluster distinctly from the phage(s) represented by the Y04AAS1 CRISPR spacers (<85% identity) derived from the Obsidian Pool complex approximately 25 km away. Though the DS metagenome is not complete, failure to find identical Y04AAS1 markers suggests the F8 Hydrogenobaculum strain per se may not inhabit DS or is in relative low abundance. Rather, the data are perhaps consistent with the interpretation that there are likely numerous Hydrogenobaculum populations in DS that genetically span a continuum between the DS and the F8 organisms considered in this study. Evidence of Sulfurihydrogenibium population-level diversity along temperature and chemical gradients in the outflow channel of the Coffee Pots geothermal feature has been reported previously (8). As well, evidence of population diversification has been demonstrated among Sulfolobus islandicus populations physically separated by similar or even shorter geographical distances in YNP (68–70) than those characterized here for DS and the Obsidian Pool complex.
In addition to apparent adaptation and or genetic drift, the evidence also suggests that roughly half of the genetic/functional differences between the DS and F8 organisms may be due to HGT (see Table S2 in the supplemental material). Nearly half of the predicted HGT genes are found in a thermophile, an acidophile, or an acidothermophile (see Table S2), providing an ecological linkage for acquisition. The kdp genes were selected to assess HGT because they encode a characterized function (potassium transport, as opposed to a hypothetical protein), and they occur in the F8 Hydrogenobaculum organism as a complete operon that, along with flanking DNA, is predicted to have derived from the same organism, a T. narugense-like bacterium (Fig. 6; see also Table S2), and associated with a phylum clearly different from the Aquificae (Fig. 6). The kdp genes used for this analysis were intentionally selected so as to represent a range of phyla, and it is noteworthy that in general the kdpA clade structure corresponded very well with 16S rRNA gene-based phyla (Fig. 6). The F8 and T. narugense kdpA genes were assigned to the phylum Firmicutes, consistent with the current placement of T. narugense in Bergey's Manual (52). However, the T. narugense 16S rRNA gene places this organism closer to the candidate phylum OP9 (53), first established as a group of environmental clones amplified from Obsidian Pool (71). Regardless, the current evidence suggests the kdp operon in the F8 strain ultimately derived from a firmicute. Perhaps it is not coincidental that T. narugense is related to organisms in Obsidian Pool, as this feature is connected to OP-P, which was in turn connected to the F8 outflow channel when Y04AAS1 was isolated (A.-L. Reysenbach, personal communication; refer to Fig. S1C in the supplemental material). A high-stringency search (≥90% match criterion) of the OP-P metagenome identified a T. narugense-like organism that coexists with Hydrogenobaculum at appreciable levels (Fig. 7), providing evidence of an ecological linkage that would allow for HGT between Hydrogenobaculum Y04AAS1 and T. narugense and contributes to an ecotype designation for Y04AAS1.
ACKNOWLEDGMENTS
This work was supported by the U.S. National Aeronautics and Space Administration (Exobiology Program NAG5-8807, NNG04GR46G), the U.S. Department of Energy Joint Genome Institute supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231, the U.S. National Science Foundation Research Coordination Network (BIO 0342269) and Microbial Observatories Program (MCB-0621291), and the Montana Agricultural Experiment Station (project 911310) to T.R.M. This work was also supported by NSF grant EAR-1123649 to E.L.S.
We thank Fred Cohan for stimulating discussions.
Footnotes
Published ahead of print 22 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03591-12.
REFERENCES
- 1. Stohr R, Waberski A, Volker H, Tindall BJ, Thomm M. 2001. Hydrogenothermus marinus gen. nov., sp. nov., a novel thermophilic hydrogen-oxidizing bacterium, recognition of Calderobacterium hydrogenophilum as a member of the genus Hydrogenobacter and proposal of the reclassification of Hydrogenobacter acidophilus as Hydrogenobaculum acidophilum gen. nov., comb. nov., in the phylum ‘Hydrogenobacter/Aquifex.’ Int. J. Syst. Evol. Microbiol. 51:1853–1862 [DOI] [PubMed] [Google Scholar]
- 2. Boyd ES, Leavitt WD, Geesey GG. 2009. CO(2) uptake and fixation by a thermoacidophilic microbial community attached to precipitated sulfur in a geothermal spring. Appl. Environ. Microbiol. 75:4289–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clingenpeel SR, D'Imperio S, Oduro H, Druschel GK, McDermott TR. 2009. Cloning and in situ expression studies of the Hydrogenobaculum arsenite oxidase genes. Appl. Environ. Microbiol. 75:3362–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Imperio S, Lehr CR, Oduro H, Druschel G, Kuhl M, McDermott TR. 2008. Relative importance of H2 and H2S as energy sources for primary production in geothermal springs. Appl. Environ. Microbiol. 74:5802–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Donahoe-Christiansen J, D'Imperio S, Jackson CR, Inskeep WP, McDermott TR. 2004. Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 70:1865–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eder W, Huber R. 2002. New isolates and physiological properties of the Aquificales and description of Thermocrinis albus sp. nov. Extremophiles 6:309–318 [DOI] [PubMed] [Google Scholar]
- 7. Ferrera I, Longhorn S, Banta AB, Liu Y, Preston D, Reysenbach AL. 2007. Diversity of 16S rRNA gene, ITS region and aclB gene of the Aquificales. Extremophiles 11:57–64 [DOI] [PubMed] [Google Scholar]
- 8. Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD. 2008. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 74:4910–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson CR, Langner HW, Donahoe-Christiansen J, Inskeep WP, McDermott TR. 2001. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3:532–542 [DOI] [PubMed] [Google Scholar]
- 10. Langner HW, Jackson CR, McDermott TR, Inskeep WP. 2001. Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ. Sci. Technol. 35:3302–3309 [DOI] [PubMed] [Google Scholar]
- 11. Mathur J, Bizzoco RW, Ellis DG, Lipson DA, Poole AW, Levine R, Kelley ST. 2007. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl. Environ. Microbiol. 73:2612–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reysenbach A-L, Banta A, Civellow S, Daly J, Mitchel K, Lalonde S, Konhauser K, Rodman A, Rustenholtz K, Takacs-Vesbach C. 2005. The Aquificales of Yellowstone National Park, p 129–142 In Inkseep WP, McDermott TR. (ed), Geothermal biology and geochemistry in Yellowstone National Park: workshop proceedings from the Thermal Biology Institute's Yellowstone National Park conference, October 2003 Montana State University, Bozeman, MT [Google Scholar]
- 13. Burgess EA, Unrine JM, Mills GL, Romanek CS, Wiegel J. 2012. Comparative geochemical and microbiological characterization of two thermal pools in the Uzon Caldera, Kamchatka, Russia. Microb. Ecol. 63:471–489 [DOI] [PubMed] [Google Scholar]
- 14. Reysenbach AL, Hamamura N, Podar M, Griffiths E, Ferreira S, Hochstein R, Heidelberg J, Johnson J, Mead D, Pohorille A, Sarmiento M, Schweighofer K, Seshadri R, Voytek MA. 2009. Complete and draft genome sequences of six members of the Aquificales. J. Bacteriol. 191:1992–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spear JR, Walker JJ, McCollom TM, Pace NR. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. U. S. A. 102:2555–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spear JR, Walker JJ, Pace NR. 2005. Hydrogen and primary productivity: inference of biogeochemistry from phylogeny in a geothermal ecosystem, p 113–127 In Inskeep WP, McDermott TR. (ed), Geothermal biology and geochemistry in Yellowstone National Park, 1st ed Montana State University Publications, Bozeman, MT [Google Scholar]
- 17. D'Imperio S, Lehr CR, Breary M, McDermott TR. 2007. Autecology of an arsenite chemolithotroph: sulfide constraints on function and distribution in a geothermal spring. Appl. Environ. Microbiol. 73:7067–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shock EL, Holland M, Meyer-Dombard D, Amend JP, Osburn GR, Fischer TP. 2010. Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, U.S.A. Geochim. Cosmochim. Acta 74:4005–4043 [Google Scholar]
- 19. Windman T, Zolotova N, Schwandner F, Shock EL. 2007. Formate as an energy source for microbial metabolism in chemosynthetic zones of hydrothermal ecosystems. Astrobiology 7:873–890 [DOI] [PubMed] [Google Scholar]
- 20. Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, McCutcheon JP, McDonald BR, Moran NA, Bristow J, Cheng JF. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314 doi:10.1371/journal.pone.0010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S, Malmstrom R, Stepanauskas R, Cheng JF. 2011. Decontamination of MDA reagents for single cell whole genome amplification. PLoS One 6:e26161 doi:10.1371/journal.pone.0026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, Jennings R, Fouke BW, Reysenbach AL, Roberto F, Young M, Schwartz A, Boyd ES, Badger JH, Mathur EJ, Ortmann AC, Bateson M, Geesey G, Frazier M. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773 doi:10.1371/journal.pone.0009773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bennett S. 2004. Solexa Ltd. Pharmacogenomics 5:433–438 [DOI] [PubMed] [Google Scholar]
- 24. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 27. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 28. Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195–202 [DOI] [PubMed] [Google Scholar]
- 29. Han C, Chain P. 2006. Finishing repeat regions automatically with Dupfinisher, p 141–146 In Arabnia HR, Valafar H. (ed), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology, CSREA Press, Las Vegas, NV, 26 to 29 June 2006 [Google Scholar]
- 30. Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278 [DOI] [PubMed] [Google Scholar]
- 31. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 33. Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jansen R, van Embden JD, Gaastra W, Schouls LM. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 6:23–33 [DOI] [PubMed] [Google Scholar]
- 37. Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81–91 [DOI] [PubMed] [Google Scholar]
- 39. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ackerman 2006. Biogeochemical gradients and energetics in geothermal systems of Yellowstone National Park. M.S. thesis. Montana State University, Bozeman, MT [Google Scholar]
- 41. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 42. Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64:475–493 [DOI] [PubMed] [Google Scholar]
- 43. Godde JS, Bickerton A. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718–729 [DOI] [PubMed] [Google Scholar]
- 44. Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60 doi:10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah SA, Garrett RA. 2011. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res. Microbiol. 162:27–38 [DOI] [PubMed] [Google Scholar]
- 49. Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 50. Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14:321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200–207 [DOI] [PubMed] [Google Scholar]
- 52. Mori K, Hanada S. 2009. Genus I. Thermodesulfobium, p 1268–1274 In Vos PD, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB. (ed), Bergey's manual of systematic bacteriology, vol 3 The Firmicutes. Springer-Verlag, New York, NY [Google Scholar]
- 53. Mori K, Kim H, Kakegawa T, Hanada S. 2003. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 7:283–290 [DOI] [PubMed] [Google Scholar]
- 54. Brock TD. 1971. Bimodal distribution of pH values of thermal springs of the world. Geol. Soc. Am. Bull. 82:1393–1394 [Google Scholar]
- 55. Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551–554 [DOI] [PubMed] [Google Scholar]
- 56. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344 doi:10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Green JL, Bohannan BJ, Whitaker RJ. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039–1043 [DOI] [PubMed] [Google Scholar]
- 58. Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 59. Wayne LG, Brenner DJ, Colwell PR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463–464 [Google Scholar]
- 60. Cohan FM. 2001. Bacterial species and speciation. Syst. Biol. 50:513–524 [DOI] [PubMed] [Google Scholar]
- 61. Freedman Z, Zhu C, Barkay T. 2012. Mercury resistance and mercuric reductase activities and expression among chemotrophic thermophilic Aquificae. Appl. Environ. Microbiol. 78:6568–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Inskeep WP, Macur RE, Hamamura N, Warelow TP, Ward SA, Santini JM. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934–943 [DOI] [PubMed] [Google Scholar]
- 63. Hamamura N, Macur RE, Korf S, Ackerman G, Taylor WP, Kozubal M, Reysenbach AL, Inskeep WP. 2009. Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 11:421–431 [DOI] [PubMed] [Google Scholar]
- 64. Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050 [DOI] [PubMed] [Google Scholar]
- 65. Rachel R, Bettstetter M, Hedlund BP, Haring M, Kessler A, Stetter KO, Prangishvili D. 2002. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch. Virol. 147:2419–2429 [DOI] [PubMed] [Google Scholar]
- 66. Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. 2008. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 74:4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Snyder JC, Wiedenheft B, Lavin M, Roberto FF, Spuhler J, Ortmann AC, Douglas T, Young M. 2007. Virus movement maintains local virus population diversity. Proc. Natl. Acad. Sci. U. S. A. 104:19102–19107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grogan DW, Ozarzak MA, Bernander R. 2008. Variation in gene content among geographically diverse Sulfolobus isolates. Environ. Microbiol. 10:137–146 [DOI] [PubMed] [Google Scholar]
- 69. Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106:8605–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
- 71. Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yarza P, Ludwig W, Euzeby J, Amann R, Schleifer KH, Glockner FO, Rossello-Mora R. 2010. Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst. Appl. Microbiol. 33:291–299 [DOI] [PubMed] [Google Scholar]
- 73. Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glockner FO, Rossello-Mora R. 2008. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250 [DOI] [PubMed] [Google Scholar]