Abstract
Listeriosis is a disease that causes significant economic losses at the farm level because of high morbidity and mortality in ruminants. This study was performed to investigate the role of ruminants in the epidemiology of listeriosis in northern Italy and the possible association of animal-adapted strains of Listeria monocytogenes with strains associated with human disease. Twenty ruminant rhombencephalitis isolates previously confirmed as L. monocytogenes by bacteriology and PCR were characterized by serotyping, pulsed-field gel electrophoresis, multi-virulence-locus sequence typing (MVLST), and multiplex single nucleotide polymorphism (mSNP) typing for the detection of epidemic clones. Subtyping results were subsequently compared with those obtained from human, food, and environmental isolates of L. monocytogenes, including 311 isolates from the University of Turin, Grugliasco, Italy, and 165 isolates representing major human listeriosis outbreaks worldwide, in addition to other unrelated isolates. Both mSNP typing and MVLST showed that 60% of the isolates analyzed belonged to epidemic clone I (ECI), which has been epidemiologically linked to several human outbreaks of listeriosis. In particular, the 1981 Canada outbreak was linked to the use of sheep manure and the 1985 California outbreak was linked to the use of raw cow's milk. In our study, ECI isolates were collected from different ruminant species on geographically and temporally distinct occasions for the last 13 years. Our results support the hypothesis that ruminants represent possible natural reservoirs of L. monocytogenes strains capable of causing epidemics of listeriosis in humans.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, facultative, saprophytic, food-borne pathogen that causes listeriosis most commonly in immunocompromised mammals (1, 2). The main clinical manifestations of listeriosis in both humans and animals include encephalitis, septicemia, and abortion (1, 3). In ruminants, listeriosis is the most important central nervous system disease in various European neuropathological surveillance programs (4). The disease is most commonly acquired through the consumption of contaminated feed and food, particularly silage by ruminants and ready-to-eat (RTE) foods by humans (1, 5–7).
In the past few years, several phenotypic and genotypic typing systems have been able to subtype L. monocytogenes at various levels of taxonomic resolution, from genetic lineages (3) to serotypes (1) to clonal complexes (8) to epidemic clones (ECs) (5, 9) to strains (5). In particular, ECs of L. monocytogenes are defined as groups of genetically related strains that have been implicated in geographically and temporally unrelated outbreaks (5, 9). Although new typing methods have been developed (10–13), pulsed-field gel electrophoresis (PFGE) is still considered the gold standard method for the subtyping of L. monocytogenes (14) and, with serotyping, has been routinely used to characterize human and food isolates (15–20). Multi-virulence-locus sequence typing (MVLST) is a sequence typing method that has shown high discriminatory power, epidemiologic concordance, stability, typeability, and reproducibility. MVLST correctly identified all seven currently known ECs of L. monocytogenes (21–23).
PFGE and other molecular subtyping techniques have rarely been used to characterize animal isolates of L. monocytogenes (24–28). In fact, most isolates of L. monocytogenes from animal clinical cases have been characterized by serotyping and ribotyping (5, 20, 29). The latter has a lower discriminatory power than other fragment-based techniques, such as PFGE (14). Serotyping is able to identify 13 serotypes, but the majority of the strains associated with human listeriosis cases and outbreaks are represented by only three of them: serotypes 1/2a, 1/2b, and 4b (14). When a study used PFGE for the characterization of animal clinical isolates of L. monocytogenes, it presented preliminary data supporting a genetic correlation between individual strains responsible for veterinary clinical cases and strains recovered from cases of human listeriosis. It was therefore hypothesized that food animals could represent reservoirs of L. monocytogenes able to cause human infections (26). The role of ruminants as possible reservoirs of L. monocytogenes strains that are pathogenic to humans was also suggested by other studies (30–32). However, those studies did not systematically compare animal clinical isolates with those associated with human outbreaks.
Listeriosis is an important issue from a veterinary standpoint, considering the significant economic losses at the farm level due to the morbidity and high mortality it causes in animals (33). Veterinary listeriosis can also pose a potential risk to food safety and public health because of a link between the farm environment and human infection (33). Studies aimed at characterizing isolates obtained from animals suffering from listeriosis are needed in order to better understand the pathophysiologic, epidemiologic, and genetic mechanisms involved in all clinical forms of the disease in different species. Additionally, further evaluation of the genetic diversity of L. monocytogenes isolates from ruminant rhombencephalitis and their degree of overlap with strains from human listeriosis cases is also desirable. To this end, L. monocytogenes isolates from ruminant rhombencephalitis cases collected over a 13-year period by the Center for Animal Encephalopathy (CEA) at the Istituto Zooprofilattico Sperimentale of Piemonte, Liguria, and Valle d'Aosta and within the Department of Veterinary Sciences of the University of Turin, Grugliasco, Italy, were characterized by serotyping, PFGE, multiplex single nucleotide polymorphism (mSNP) typing, and MVLST. Typed strains were compared with different sets of isolates collected from human outbreaks and sporadic cases and food and environmental sources previously characterized by PFGE (34) and MVLST (13, 21, 22). Molecular subtyping results were subsequently compared with microscopic brain lesions in order to determine possible associations between the severity and distribution of brain lesions and the location, animal species, and subtypes of L. monocytogenes strains isolated.
MATERIALS AND METHODS
Twenty cases of ruminant rhombencephalitis (12 cattle, five sheep, and three goats; Table 1) were diagnosed within the CEA and the Department of Veterinary Sciences, University of Turin, Grugliasco, Italy. The animals either died spontaneously or were sacrificed because of neurological signs and a poor prognosis. Brains were sampled and divided by an aseptic paramedian cut, and the larger portion was fixed in 10% buffered formaldehyde solution; the smaller portion was frozen at −20°C. Because of neurologic disease, all cases were also submitted for confirmatory tests for transmissible spongiform encephalopathies (TSE) (35).
Table 1.
Description of the 20 L. monocytogenes isolates obtained from animal clinical cases
| ID | Source | Yr isolated, location | Serotype | EC | AscI pulsotype | ApaI pulsotype | Combined profile | MVLST VT |
|---|---|---|---|---|---|---|---|---|
| 01 | Bovine | 2005, Lombardy | 4b | ECI | S1 | A3 | P1 | 20 |
| 02 | Bovine | 2005, Lombardy | 1/2b | —a | Uniqueb | A2 | Unique | 78 |
| 03 | Bovine | 2007, Piedmont | 4e | — | S4 | A4 | P4 | 75 |
| 04 | Caprine | 2009, Piedmont | 1/2a | — | S5 | Unique | Unique | 76 |
| 05 | Ovine | 2001, Aosta Valley | 4b | ECI | S1 | Unique | Unique | 20 |
| 06 | Bovine | 2001, Piedmont | 4b | ECI | S1 | A3 | P1 | 20 |
| 07 | Bovine | 2001, Piedmont | 4b | ECI | S2 | A2 | P2 | 20 |
| 08 | Bovine | 2001, Lombardy | 4b | ECI | S2 | A2 | P2 | 20 |
| 09 | Bovine | 2002, Lazio | 4e | ECI | S3 | A1 | P3 | 20 |
| 10 | Bovine | 2002, Piedmont | 4b | ECI | S2 | A2 | P2 | 20 |
| 11 | Bovine | 2002, Emilia Romagna | 4b | — | Unique | A4 | Unique | 75 |
| 12 | Bovine | 2003, Veneto | 4b | ECI | S3 | A1 | P3 | 20 |
| 13 | Bovine | 2009, Piedmont | 4e | ECI | Unique | Unique | Unique | 20 |
| 14 | Ovine | 2010, Piedmont | 4b | — | S4 | A4 | P4 | 75 |
| 15 | Bovine | 2010, Piedmont | 4b | ECI | S3 | A1 | P3 | 20 |
| 16 | Ovine | 2009, Piedmont | 1/2b | — | Unique | Unique | Unique | 08 |
| 17 | Ovine | 1999, Aosta Valley | 4b | ECI | S1 | Unique | Unique | 20 |
| 18 | Caprine | 2000, Piedmont | 4b | ECI | S3 | A1 | P3 | 20 |
| 19 | Ovine | 2002, Liguria | 1/2a | — | S5 | Unique | Unique | 76 |
| 20 | Caprine | 2011, Piedmont | 1/2a | — | Unique | Unique | Unique | 77 |
—, not an epidemic clone.
Unique pulsotype not similar to any other observed in the database.
Histopathologic and IHC evaluations.
In each case, the obex; pons; mesencephalon; cerebellum; hippocampus; thalamus; basal nuclei; and occipital, parietal, and frontal cortex were trimmed. Subsequently, the tissues were routinely processed and embedded in paraffin and 4-μm tissue sections were cut for histopathologic and immunohistochemical (IHC) evaluations. The sections used for histopathologic evaluation were stained with hematoxylin and eosin (H&E), histopathologic lesions were categorized by semiquantitative assessment of each anatomic brain region for the extent and size of microabscesses/perivascular cuffs by a previously established approach (36), and semiquantitative scores of 0 to 4 were assigned. IHC was performed by using the following protocol. For antigen retrieval, paraffin-included 4-μm brain sections were treated with citrate buffer (pH 6.0) at 100°C for 15 min. Subsequently, the sections were exposed to primary antibody (polyclonal anti-Listeria species antibody, code 43251; 1:250 dilution, Virostat, Portland, OR) and incubated overnight. The sections were then exposed to a secondary antibody and immunoperoxidase stained with the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA). Microscopically positive antigen-antibody reactions were visualized after incubation with the chromogen 3,3′-diaminobenzidine (DAB) for 1 min at room temperature. Phosphate-buffered saline was used to wash the slides between the different preparation steps. H&E-stained and IHC slides were examined with a Leica DM LS2 light microscopic (Leica Microsystems, Wetzlar, Germany) at different magnifications. H&E-stained slides were evaluated at magnification levels of ×25, ×100, ×200, and ×400; a magnification of ×400 was used to evaluate IHC immunopositivity.
Bacteriology, serotyping, and DNA isolation.
The frozen brain samples were thawed, placed in liquid medium (Demi-Fraser broth and Fraser broth; Microbiol, Bagno a Ripoli, Italy), and incubated at 37°C for 48 h and at 4°C for 10 days. Subsequently, cultures were streaked onto Listeria Oxford agar (Microbiol) and incubated at 37°C for 48 h. Suspected Listeria species colonies were confirmed as L. monocytogenes with a miniaturized API System (bioMérieux, Marcy l'Etoile, France) and a colorimetric Vitek 2 identification System (bioMérieux, Bagno a Ripoli, Italy). All L. monocytogenes animal isolates were serotyped with antisera against the O and H antigens according to the manufacturer's instructions (Denka Seiken Co., Ltd., Tokyo, Japan). Serotypes were further confirmed with a multiplex PCR (37).
All isolates were stored at −80°C in cryogenic vials. Prior to DNA extraction, isolates were grown overnight at 37°C in tryptone soy broth (TSB; Acumedia, Lansing, MI). DNA was extracted by boiling with the following protocol. One milliliter of TSB culture (108 CFU/ml) was centrifuged at 12,000 × g for 5 min; the pellet was subsequently resuspended in 1 ml of phosphate-buffered saline and boiled for 5 min. After centrifugation, the supernatant was stored at −20°C until use. DNA was quantified with a Bio-Photometer 6131 spectrophotometer (Eppendorf AG, Hamburg, Germany) and diluted to a concentration of 50 to 60 ng/μl prior to PCR.
Duplex PCR, mSNP typing, PFGE, and MVLST.
Isolates were confirmed as L. monocytogenes by a duplex PCR-minisequencing assay able to identify L. monocytogenes and five other Listeria species (38). Ruminant isolates were also analyzed with an mSNP typing screening assay able to identify five ECs (ECI, ECII, ECIII, ECIV, and ECV) of L. monocytogenes (39).
PFGE was performed by including genomic DNA in agarose plugs prior to digestion with the AscI and ApaI restriction enzymes (New England BioLabs), followed by PFGE with a Chef DR III system (Bio-Rad, Hercules, CA) (40), and pulsotypes were analyzed as described elsewhere (34). Isolates were considered to belong to the same cluster when they showed a PFGE similarity level (SL) of at least 80%, while isolates with an SL of 100% were considered to be identical. PFGE profiles were subsequently compared with a set of 311 well-characterized L. monocytogenes strains isolated from environmental, food, and human clinical samples (34).
Intragenic regions of six virulence genes (clpP, dal, inlB, inlC, lisR, and prfA) were amplified, resolved, purified, and sequenced as previously described (21). Both forward and reverse strands were sequenced, and gene sequences were subsequently concatenated with Sequence Matrix 1.7.8 (41) and compared with sequences from 165 isolates representing major listeriosis outbreaks worldwide (13, 21, 22) with MEGA v5.0 (42). New allelic sequences (with at least one nucleotide difference) were assigned arbitrary virulence type (VT) numbers as previously described (23). The neighbor-joining method in MEGA 5.0 was used to construct an unrooted neighbor-joining tree by using the number of nucleotide differences in the concatenated sequences of six loci with 1,000 bootstrap tests.
Statistical analysis.
Brain lesion scores (microabscesses and perivascular cuffing lesion scores) were compared with geographic origins, ruminant species, and L. monocytogenes serotypes by using Kruskal-Wallis and Wilcoxon rank sum tests. Correlations between microabscesses and perivascular cuffing lesion scores were estimated with a Spearman rank correlation test. Test results were considered statistically significant when the P value was <0.05. All statistical analyses were conducted with R statistical software (43).
Nucleotide sequence accession numbers.
The gene sequences determined in this study were deposited in the GenBank database under accession numbers JX944009.
RESULTS
Histopathology and immunohistochemistry.
No macroscopic lesions were observed in any of the cases analyzed in this study. However, microscopic brain lesions were observed and included mainly multifocal microabscesses, perivascular cuffing, and edema. Microabscesses (Fig. 1) were characterized by multifocal randomly distributed aggregates of neutrophils, small-to-coalescing foci of liquefactive necrosis, and edema. Microabscesses were observed in both gray matter and white matter but were more frequently observed in the white matter of rostral areas (cerebellum, mesencephalon, and thalamus) than in the brainstem. Multifocal gliosis, diffuse infiltration of neutrophils and microglial cells, and neuronal necrosis were observed in the areas adjacent to microabscesses. The latter particularly involved the brainstem and cerebellum (data not shown). The rostral areas of the brain (thalamus, basal nuclei, and adjacent parietal cortex) were involved only when extensive microscopic lesions were present in the brainstem. Perivascular cuffings (Fig. 1A) were observed in all cases and were composed of at least one layer of inflammatory cells, most frequently lymphocytes and sporadically macrophages. In two cases, neutrophils predominated in the perivascular cuffings. The emergences of cranial nerves (mostly trigeminal and facial nerves) in the brainstem were affected by mostly microabscesses. All cases were positive by immunohistochemistry, thus confirming that Listeria species bacteria were present and associated with brain lesions. Immunopositivity was frequently observed in the cytoplasm of neutrophils in the microabscesses, in macrophages surrounding microabscesses (Fig. 1D), and in the neurophils but less frequently in the perivascular cuffings and rarely in the cytoplasm of neurons. Immunopositivity was detected in the obex in all of the animals, followed by the brainstem (67% of the bovines and 100% of the small ruminants) and mesencephalon (50% of the bovines and 75% of the small ruminants). Confirmatory tests for TSEs were negative in all cases.
Fig 1.
Histopathologic evaluations. (A) Bovine cerebellum. Multifocal-to-coalescing microabscesses (arrows) and mononuclear perivascular cuffing (asterisks) in the white matter. H&E staining. Magnification, ×25. (B) Ovine brainstem. Malacic area (arrows) composed mainly of neutrophils. H&E staining. Magnification, ×25. (C) Bovine brainstem. Microabscess composed mainly of macrophages and a few neutrophils. H&E staining. Magnification, ×400. (D) Caprine brainstem. Immunopositivity (brown areas) for Listeria species in the cytoplasm of macrophages and neutrophils within a microabscess. DAB chromogen counterstained with hematoxylin. Magnification, ×400.
Bacteriology, serotyping, and EC screening.
All isolates were confirmed as L. monocytogenes. Numbers of cases, animal species, serotypes, sources, and molecular subtyping results are summarized in Table 1. Serotype 4b was detected in 12 cases (60%; 8 bovine, 3 ovine, and 1 caprine), serotype 4e was detected in 3 cases (15%; all bovine), serotype 1/2a was detected in 3 cases (15%; 1 ovine and 2 caprine), and serotype 1/2b was detected in 2 cases (10%; 1 ovine and 1 bovine). The mSNP typing screening assay showed the ECI-specific profile in 12 isolates (60%).
PFGE profiles and MVLST VTs of the 20 animal isolates.
Thirteen combined PFGE profiles and six MVLST VTs were identified in the 20 L. monocytogenes isolates analyzed here (Fig. 2 and Table 1). PFGE distinguished cluster I grouping 13 isolates (SL = 82.3%) and cluster II grouping 3 isolates (SL = 96.5%). Four isolates (no. 02, 16, 19, and 20) had an SL of 65.6% to all of the other isolates. Isolates were partially divided according to serotypes (Fig. 2).
Fig 2.
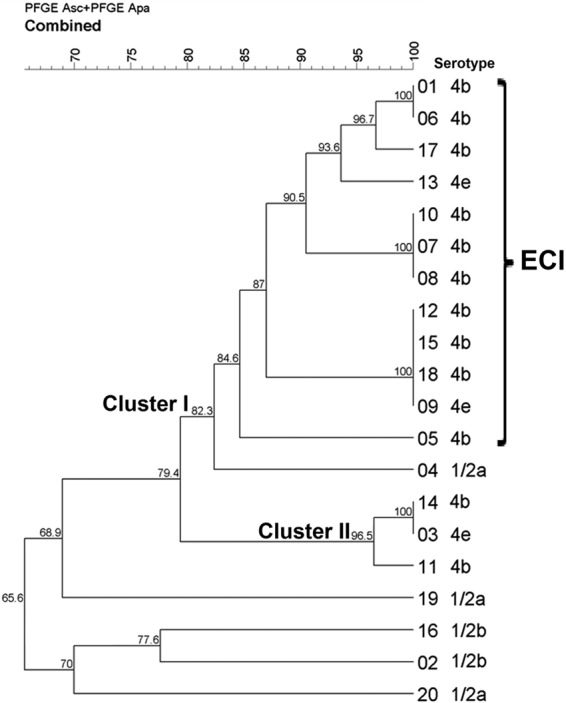
Dendrogram produced by the unweighted-pair group method using average linkages showing clustering by Dice coefficient of the combined PFGE (AscI and ApaI) profiles of the 20 L. monocytogenes isolates obtained from animal cases analyzed in this study. PFGE divided the L. monocytogenes animal isolates into nine unique and four shared profiles, P1 (two isolates), P2 (three isolates), P3 (four isolates), and P4 (two isolates).
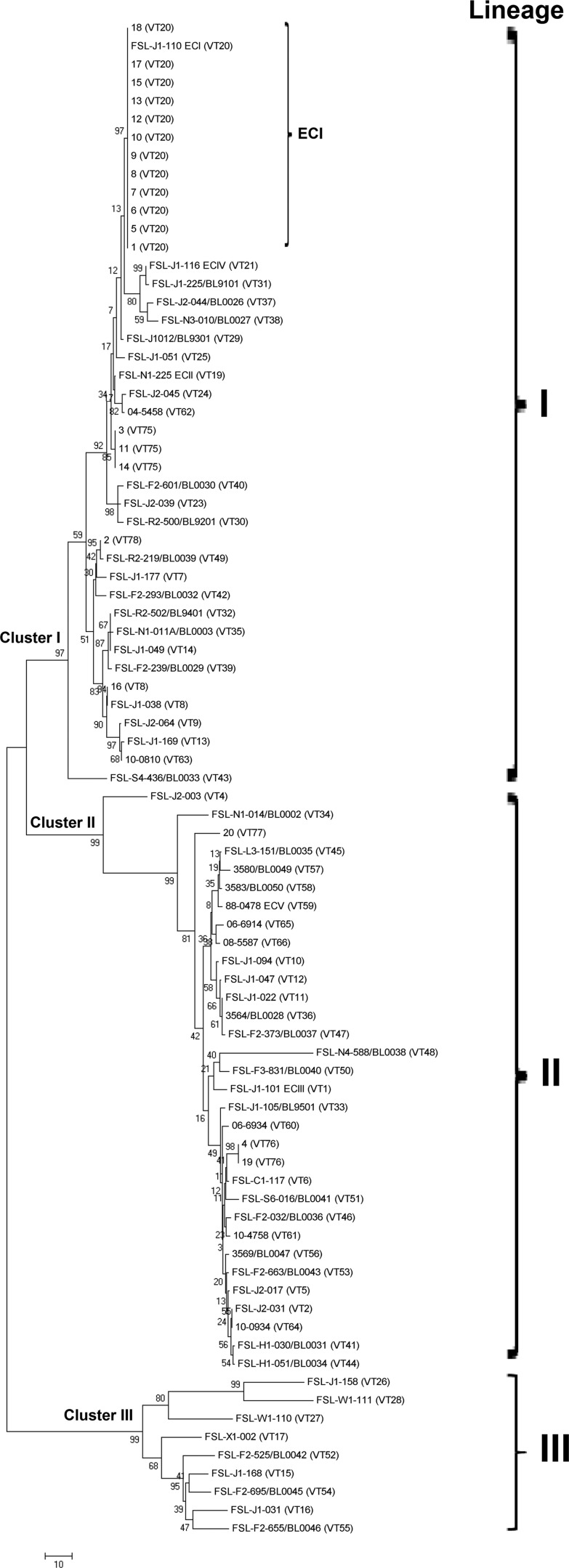
A total concatenation length of 2,606 bp was observed for the six virulence genes analyzed by MVLST. New VTs were assigned for isolates 3, 11, and 14 (VT75); 4 and 19 (VT76); 20 (VT77); and 2 (VT78). Overall, MVLST identified three shared VTs (VT20, VT75, and VT76, composed of 12, 2, and 3 isolates, respectively) and three unique ones (VT8, VT77, and VT78). The MVLST neighbor-joining tree revealed three main clusters supported by bootstrap values of 92, 98, and 100%, respectively, clearly dividing isolates according to serotype. The 12 isolates presumptively identified as ECI by mSNP typing were confirmed as ECI (VT20) by MVLST. These 12 VT20 isolates showed four shared PFGE profiles and one unique profile (Table 1) and grouped at an 84.6% SL into cluster I. The VT75 isolates showed two shared profiles and one unique profile and grouped at a 96.5% SL into cluster II, whereas VT76 showed two unique profiles and grouped at a 68.9% SL.
Comparison with previously subtyped strains.
None of the 20 L. monocytogenes ruminant clinical isolates had the same PFGE profile as any other sample in our PFGE database (data not shown). ECI isolates 9, 12, 15, and 18 had an SL of 90.1% with human clinical isolate U1, also previously identified as ECI (39).
The MVLST neighbor-joining tree revealed three main clusters that clearly separated serotypes, supported by bootstrap values of 97, 99, and 99%. Overall, clusters I, II, and III corresponded to lineages I, II, and III, respectively (Fig. 3). Only one isolate other than those belonging to VT20 showed a VT previously observed in human clinical cases, as isolate 16 (cluster I) had the same VT (VT8) as an L. monocytogenes strain of serotype 1/2b responsible for a human sporadic clinical case (13). VT78 differed by one SNP from VT49, observed in a food environmental isolate (21), and VT76 differed by three SNPs from a human clinical isolate assigned to VT6 (13). Isolate 20 was not closely related to any other isolate included in the comparison.
Fig 3.
Unrooted neighbor-joining tree computed in MEGA 5.0 (52) based on the comparison of MVLST results for the L. monocytogenes ruminant isolates investigated in this study (20 clinical veterinary isolates) with 165 isolates previously characterized by MVLST (13, 21, 22). Statistical support was provided by bootstrapping with 1,000 replicates. The scale bar corresponds to the number of differences observed over the alignment of 2,606 bp.
Statistical analysis.
No significant associations between lesions, ruminant species, geographic origins, and L. monocytogenes serotypes were found (Kruskal-Wallis rank sum test; P > 0.05 in all cases). A significant positive correlation between the grade of microabscesses and perivascular cuffing was observed (Spearman rank correlation rho = 0.5702; P < 0.01), particularly when considering only bovines (Spearman rank correlation rho = 0.8694; P < 0.001). However, no significant correlation was detected when considering only small ruminants (Spearman rank correlation rho = 0.1443; P = 0.733).
DISCUSSION
Based on the type and severity of histologic lesions and the statistical significance of more acute meningoencephalitis and rostral spread of lesions, small ruminants appear to be less capable than cattle of eliminating L. monocytogenes from their brains and thus are more susceptible to listeriosis once the pathogen reaches the brain, which is in line with previous observations (1, 4, 33). Our findings support the hypothesis of pathogenesis of L. monocytogenes migration along the cranial nerves through the brains of mammals (4, 33, 44–46). However, the pathogenic mechanisms by which bacteria cause neuron injury by centripetally migrating into the brains of both humans and animals are still poorly understood (2, 46, 47) and thus require further investigation.
The prevalence of serotypes corresponding to lineage I reported in this study clearly supports this lineage as highly common in mammals (1, 3, 6, 9, 17, 19, 28, 30, 48). Lineage I has been frequently linked to human cases and associated with the encephalitic form of the disease in ruminants (6, 30, 49). Lineage II has been described as genetically more diverse and more frequently associated with both encephalitic and septicemic forms of listeriosis in ruminants (6, 49). Although reported to cause up to 10% of animal listeriosis cases (49), ruminant isolates belonging to lineage III were not identified in this study, as also previously observed (30). However, in both studies, no isolates obtained from other clinical forms of animal listeriosis (e.g., septicemia and/or abortion) were included.
PFGE is currently the gold standard method of characterizing L. monocytogenes, and it is still essential in epidemiologic investigations of listeriosis outbreaks and in the subtyping of food-borne pathogens (14, 50). The strains identified here as ECI showed four different PFGE profiles. Furthermore, isolates 3, 11, and 14 had the same MVLST VT (VT75), as did isolates 4 and 19 (VT76), but showed different PFGE profiles. It has been shown that epidemiologically linked L. monocytogenes strains isolated from the same outbreak can have different PFGE profiles (22, 51–54).
An MVLST approach was used in this study in addition to PFGE, as the former allows the clustering of L. monocytogenes strains with high epidemiologic concordance and discriminatory power (13, 21). Interestingly, ECI isolates were collected from different species at different places and at different times in northern Italy, suggesting that ECI isolates have been circulating in ruminants in the same territory for at least the last 13 years. Because of the absence of surveillance programs for ruminant listeric rhombencephalitis in Italy, farmers and veterinarians are not required to report the disease, thus potentially limiting the number of diagnosed cases.
A literature review was performed to assess the prevalence of ECI in samples from different sources. The ECI-associated ribotype was found in 10.5 to 12.8% of human sporadic cases (55, 56) and in 11.8% of animal cases (56). A recent study from Portugal found a prevalence of ECI markers in 13.58% of the analyzed L. monocytogenes isolates obtained from fermented meat sausages, processing plant environments, and other raw products (16). In Italy, ECI markers were found in 17.6% of the human clinical isolates obtained between 1994 and 2007, 18% of the food isolates, and 20% of the environmental isolates (48). ECI was also detected in one out of five human sporadic clinical strains originating in Piedmont from 2002 to 2006 (39). To further support our findings, 56 food and environmental strains of serotype 4b/4e selected from the set of 311 strains and isolated from a geographic area partially matching that of the veterinary clinical ruminant isolates analyzed here were screened by PCR for the presence of the ECI-specific marker (57). Seventeen isolates (30.3%) were identified as putative ECI strains (data not shown). When the prevalence of ECI strains in ruminant clinical cases (60%) was compared to the observed prevalence in various food types (30.3%), the results of the chi-square test showed a significant difference between the percentages: chi square = 4.3; P = 0.03. Therefore, we believe that the observed high prevalence of ECI is significant, especially considering the limited number of animals affected in such a long time period and in such a large geographic area.
Overall, our MVLST results suggest that L. monocytogenes strains isolated from ruminants with rhombencephalitis constitute a genetically homogeneous group related to human, food, and environmental isolates, as also previously highlighted by other authors (6, 49, 55). This is in contrast to what was observed in 183 L. monocytogenes isolates obtained mostly from ruminants and to a lesser extent from human clinical rhombencephalitis cases, food, and the environment analyzed with multilocus variable-number tandem-repeat analysis (MLVA) (30). Their findings revealed that L. monocytogenes isolates clustered according to serotype and whether they were obtained from clinical cases or from food/environmental sources. No matches between food isolates and ruminant isolates were observed; however, some human clinical isolates and ruminant rhombencephalitis isolates had identical MLVA types, indicating ruminants as possible reservoirs for human infection, as also previously suggested (26, 31, 58). The role of ruminants as possible reservoirs for human infection is also supported by our results and the comparison of 165 isolates (from both food and clinical cases) representing the major food-borne outbreaks of listeriosis reported worldwide in the last 40 years, in addition to other unrelated isolates from sporadic cases.
EC strains may possess a greater ability to persist in the environment, survive and replicate in foods, and be transmitted to animals and humans (9). For instance, dairy farms are more frequently contaminated with L. monocytogenes than are other environments (33). The amplification and spread of L. monocytogenes into the farm environment can be attributed to ruminants via shedding of the pathogen through feces and/or contamination of raw milk due to shedding or exogenous contamination from the environment (33). In particular, ECI has been epidemiologically associated with cross contamination (e.g., the use of sheep manure as fertilizer in the 1981 Canada outbreak), the use of raw materials (outbreaks in France in 1992 and California in 1985), or postpasteurization contamination (outbreak in Switzerland from 1983 to 1987) (9). Consequently, ECs that have been causing listeriosis in animals and humans (such as ECI) may live as saprophytes in the farm environment, and raw animal foods can then play a role in the contamination of RTE foods because of cross contamination and thus may result in sporadic cases and outbreaks.
This study compared the subtypes of L. monocytogenes isolates from clinically affected ruminants to those of a significant number of clinical and food isolates obtained from the major human outbreaks of listeriosis. Our findings suggest that ruminants represent an important natural reservoir of L. monocytogenes and a potential source of strains pathogenic to humans. Additional molecular subtyping data on a larger number of L. monocytogenes isolates from farm animals with and without neurological disease and from different geographical areas/continents are needed to confirm the listeriosis link between ruminants and humans. Such data may help in the development of intervention strategies for preventing listeriosis, thus diminishing economic losses for the food and livestock industries and enhancing food safety.
ACKNOWLEDGMENTS
P. R. D. Rocha is the recipient of a Ph.D. scholarship from the Italian Ministry of Foreign Affairs. This study was financed by the Italian Health Ministry (Ministero della Salute, Finanziamento CEA 2010, 10TSE).
We acknowledge Luigi Bertolotti and Daniele Nucera for helping with statistical analysis. We also acknowledge Alessandra Sereno, Lucia Florio, and Tiziana Avanzato for technical assistance.
Footnotes
Published ahead of print 1 March 2013
REFERENCES
- 1. Low JC, Donachie W. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153: 9– 29 [DOI] [PubMed] [Google Scholar]
- 2. Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14: 584– 640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301: 79– 96 [DOI] [PubMed] [Google Scholar]
- 4. Di Palma S, Brunetti B, Doherr MG, Forster U, Hilbe M, Zurbriggen A, Vandevelde M, Oevermann A. 2012. Comparative spatiotemporal analysis of the intrathecal immune response in natural listeric rhombencephalitis of cattle and small ruminants. Comp. Immunol. Microbiol. Infect. Dis. 35: 429– 441 [DOI] [PubMed] [Google Scholar]
- 5. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65: 1811– 1829 [DOI] [PubMed] [Google Scholar]
- 6. Pohl MA, Wiedmann M, Nightingale KK. 2006. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am. J. Vet. Res. 67: 616– 626 [DOI] [PubMed] [Google Scholar]
- 7. Wiedmann M, Czajka J, Bsat N, Bodis M, Smith MC, Divers TJ, Batt CA. 1994. Diagnosis and epidemiological association of Listeria monocytogenes strains in two outbreaks of listerial encephalitis in small ruminants. J. Clin. Microbiol. 32: 991– 996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ragon M, Wirth T, Holland F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4: e1000146 doi:10.1371/journal.ppat.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng Y, Siletzky R, Kathariou S. 2008. Genomic divisions/lineages, epidemic clones, and population structure, p 337– 357 In Liu D. (ed), Handbook of Listeria monocytogenes. CRC Press, Boca Raton, FL [Google Scholar]
- 10. Laksanalamai P, Jackson SA, Mammel MK, Datta AR. 2012. High density microarray analysis reveals new insights into genetic footprints of Listeria monocytogenes strains involved in listeriosis outbreaks. PLoS One 7: e32896 doi:10.1371/journal.pone.0032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy M, Corcoran D, Buckley JF, O'Mahony M, Whyte P, Fanning S. 2007. Development and application of multiple-locus variable number of tandem repeat analysis (MLVA) to subtype a collection of Listeria monocytogenes. Int. J. Food Microbiol. 115:187– 194 [DOI] [PubMed] [Google Scholar]
- 12. Salcedo C, Arreaza L, Alcalá B, de la Fuente L, Vázquez JA. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41: 757– 762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang W, Jayarao BM, Knabel SJ. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70: 913– 920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y, Knabel SJ. 2008. Strain typing, p 203–240 In Liu D. (ed), Handbook of Listeria monocytogenes. CRC Press, Boca Raton, FL [Google Scholar]
- 15. Almeida G, Morvan A, Magalhães R, Santos I, Hogg T, Leclercq A, Teixeira P. 2010. Distribution and characterization of Listeria monocytogenes clinical isolates in Portugal, 1994-2007. Eur. J. Clin. Microbiol. Infect. Dis. 29: 1219– 1227 [DOI] [PubMed] [Google Scholar]
- 16. Ferreira V, Barbosa J, Stasiewicz M, Vongkamjan K, Moreno Switt A, Hogg T, Gibbs P, Teixeira P, Wiedmann M. 2011. Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl. Environ. Microbiol. 77: 2701– 2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gianfranceschi MV, D'Ottavio MC, Gattuso A, Bella A, Aureli P. 2009. Distribution of serotypes and pulsotypes of Listeria monocytogenes from human, food and environmental isolates (Italy 2002-2005). Food Microbiol. 26: 520– 526 [DOI] [PubMed] [Google Scholar]
- 18. Laksanalamai P, Joseph LA, Silk BJ, Burall LS, Tarr CL, Gerner-Smidt P, Datta AR. 2012. Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One 7: e42448 doi:10.1371/journal.pone.0042448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mammina C, Aleo A, Romani C, Pellissier N, Nicoletti P, Pecile P, Nastasi A, Pontello MM. 2009. Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 47:2925– 2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nappi R, Bozzetta E, Serra R, Grattarola C, Decastelli L, Florio C, Caramelli M. 2005. Molecular characterization of Listeria monocytogenes strains associated with outbreaks of listeriosis in humans and ruminants and food products by serotyping and automated ribotyping. Vet. Res. Commun. 29(Suppl 2):249–252 [DOI] [PubMed] [Google Scholar]
- 21. Chen Y, Zhang W, Knabel SJ. 2007. Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45: 835– 846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knabel S, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon N, Gilmour M. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988-2010. J. Clin. Microbiol. 50: 1748– 1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lomonaco S, Verghese B, Gerner-Smidt P, Tarr C, Gladney L, Joseph L, Katz L, Turnsek M, Frace M, Chen Y, Brow E, Meinersmann R, Berrang M, Knabel S. 2013. Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg. Infect. Dis. 19: 147– 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borucki MK, Gay CC, Reynolds J, McElwain KL, Kim SH, Call DR, Knowles DP. 2005. Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm. Appl. Environ. Microbiol. 71: 5893– 5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bundrant BN, Hutchins T, den Bakker HC, Fortes E, Wiedmann M. 2011. Listeriosis outbreak in dairy cattle caused by an unusual Listeria monocytogenes serotype 4b strain. J. Vet. Diagn. Invest. 23: 155– 158 [DOI] [PubMed] [Google Scholar]
- 26. Okwumabua O, O'Connor M, Shull E, Strelow K, Hamacher M, Kurzynski T, Warshauer D. 2005. Characterization of Listeria monocytogenes isolates from food animal clinical cases: PFGE pattern similarity to strains from human listeriosis cases. FEMS Microbiol. Lett. 249: 275– 281 [DOI] [PubMed] [Google Scholar]
- 27. Wagner M, Melzner D, Bagò Z, Winter P, Egerbacher M, Schilcher F, Zangana A, Schoder D. 2005. Outbreak of clinical listeriosis in sheep: evaluation from possible contamination routes from feed to raw produce and humans. J. Vet. Med. B Infect. Dis. Vet. Public Health 52: 278– 283 [DOI] [PubMed] [Google Scholar]
- 28. Wesley IV, Larson DJ, Harmon KM, Luchansky JB, Schwartz AR. 2002. A case report of sporadic ovine listerial menigoencephalitis in Iowa with an overview of livestock and human cases. J. Vet. Diagn. Invest. 14: 314– 321 [DOI] [PubMed] [Google Scholar]
- 29. Wiedmann M, Mobini S, Cole JR, Jr, Watson CK, Jeffers GT, Boor KJ. 1999. Molecular investigation of a listeriosis outbreak in goats caused by an unusual strain of Listeria monocytogenes. J. Am. Vet. Med. Assoc. 215: 369– 371 [PubMed] [Google Scholar]
- 30. Balandyté L, Brodard I, Frey J, Oevermann A, Abril C. 2011. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to a multilocus variable-number tandem-repeat analysis complex. Appl. Environ. Microbiol. 77: 8325– 8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boerlin P, Piffaretti JC. 1991. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 57: 1624– 1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foerster C, Vidal L, Troncoso M, Figueroa G. 2012. Characterization of Listeria monocytogenes isolates from cattle and ground beef by pulsed-field gel electrophoresis. Rev. Argent. Microbiol. 44: 195– 200 [PubMed] [Google Scholar]
- 33. Oevermann A, Zurbriggen A, Vandeveld ME. 2010. Rhombencephalitis caused by Listeria monocytogenes in humans and ruminants: a zoonosis on the rise? Interdiscip. Perspect. Infect. Dis. 2010: 632513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nucera D, Lomonaco S, Bianchi DM, Decastelli L, Grassi MA, Bottero MT, Civera T. 2010. A five year surveillance report on PFGE types of Listeria monocytogenes isolated in Italy from food and food related environments. Int. J. Food Microbiol. 140:271– 276 [DOI] [PubMed] [Google Scholar]
- 35. European Parliament 2001. Regulation (EC) No 999/2001/EC of the European Parliament and of the Council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. Official J. Eur. Communities 44: L147/1– L147/40 http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2001:147:0001:0040:EN:PDF [Google Scholar]
- 36. Oevermann A, Di Palma S, Doherr MG, Abril C, Zurbriggen A, Vandevelde M. 2010. Neuropathogenesis of naturally occurring encephalitis caused by Listeria monocytogenes in ruminants. Brain Pathol. 20: 378– 390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42: 3819– 3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dalmasso A, Rantsiou K, Cocolin L, Bottero MT. 2010. Development of a biomolecular assay for the identification of Listeria at species level. Foodborne Pathog. Dis. 7: 565– 571 [DOI] [PubMed] [Google Scholar]
- 39. Lomonaco S, Knabel SJ, Dalmasso A, Civera T, Bottero MT. 2011. Novel multiplex single nucleotide polymorphism-based method for identifying epidemic clones of Listeria monocytogenes. Appl. Environ. Microbiol. 77: 6290– 6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Graves LM, Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65: 55– 62 [DOI] [PubMed] [Google Scholar]
- 41. Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171– 180 [DOI] [PubMed] [Google Scholar]
- 42. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731– 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. R Development Core Team 2005. R: a language and environment for statistical computing, reference index version 2.2.1. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 44. Antal EA, Løberg EM, Dietrichs E, Maehlen J. 2005. Neuropathological findings in 9 cases of Listeria monocytogenes brain stem encephalitis. Brain Pathol. 15: 187– 191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barlow RM, McGorum B. 1985. Ovine listerial encephalitis: analysis, hypothesis and synthesis. Vet. Rec. 116: 233– 236 [DOI] [PubMed] [Google Scholar]
- 46. Clauss HE, Lorber B. 2008. Central nervous system infection with Listeria monocytogenes. Curr. Infect. Dis. Rep. 10: 300– 306 [DOI] [PubMed] [Google Scholar]
- 47. Drevets DA, Bronze MS. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 53: 151– 165 [DOI] [PubMed] [Google Scholar]
- 48. Franciosa G, Scalfaro C, Maugliani A, Floridi F, Gattuso A, Hodzic S, Aureli P. 2007. Distribution of epidemic clonal genetic markers among Listeria monocytogenes 4b isolates. J. Food Prot. 70: 574– 581 [DOI] [PubMed] [Google Scholar]
- 49. Jeffers GT, Bruce JL, McDonough PL, Scarlett J, Boor KJ, Wiedmann M. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147: 1095– 1104 [DOI] [PubMed] [Google Scholar]
- 50. Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytiä-Trees E, Ribot EM, Swaminathan B. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3: 9– 19 [DOI] [PubMed] [Google Scholar]
- 51. Graves LM, Hunter SB, Ong AR, Schoonmaker-Bopp D, Hise K, Kornstein L, DeWitt WE, Hayes PS, Dunne E, Mead P, Swaminathan B. 2005. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 43: 2350– 2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Centers for Disease Control and Prevention 2011. Multistate outbreak of listeriosis associated with Jensen Farms cantaloupe—United States, August-September 2011. MMWR Morb. Mortal. Wkly. Rep. 60: 1357– 1358 [PubMed] [Google Scholar]
- 53. Olsen SJ, Patrick M, Hunter SB, Reddy V, Kornstein L, MacKenzie WR, Lane K, Bidol S, Stoltman GA, Frye DM, Lee I, Hurd S, Jones TF, LaPorte TN, Dewitt W, Graves L, Wiedmann M, Schoonmaker-Bopp DJ, Huang AJ, Vincent C, Bugenhagen A, Corby J, Carloni ER, Holcomb ME, Woron RF, Zansky SM, Dowdle G, Smith F, Ahrabi-Fard S, Ong AR, Tucker N, Hynes NA, Mead P. 2005. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin. Infect. Dis. 40: 962– 967 [DOI] [PubMed] [Google Scholar]
- 54. Sauders BD, Schukken Y, Kornstein L, Reddy V, Bannerman T, Salehi E, Dumas N, Anderson BJ, Massey JP, Wiedmann M. 2006. Molecular epidemiology and cluster analysis of human listeriosis cases in three U.S. states. J. Food Prot. 69: 1680– 1689 [DOI] [PubMed] [Google Scholar]
- 55. Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70: 5833– 5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, Selander RK, Rocourt J. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 86: 3818– 3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen Y, Knabel SJ. 2007. Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73: 6299– 6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70: 4458– 4467 [DOI] [PMC free article] [PubMed] [Google Scholar]