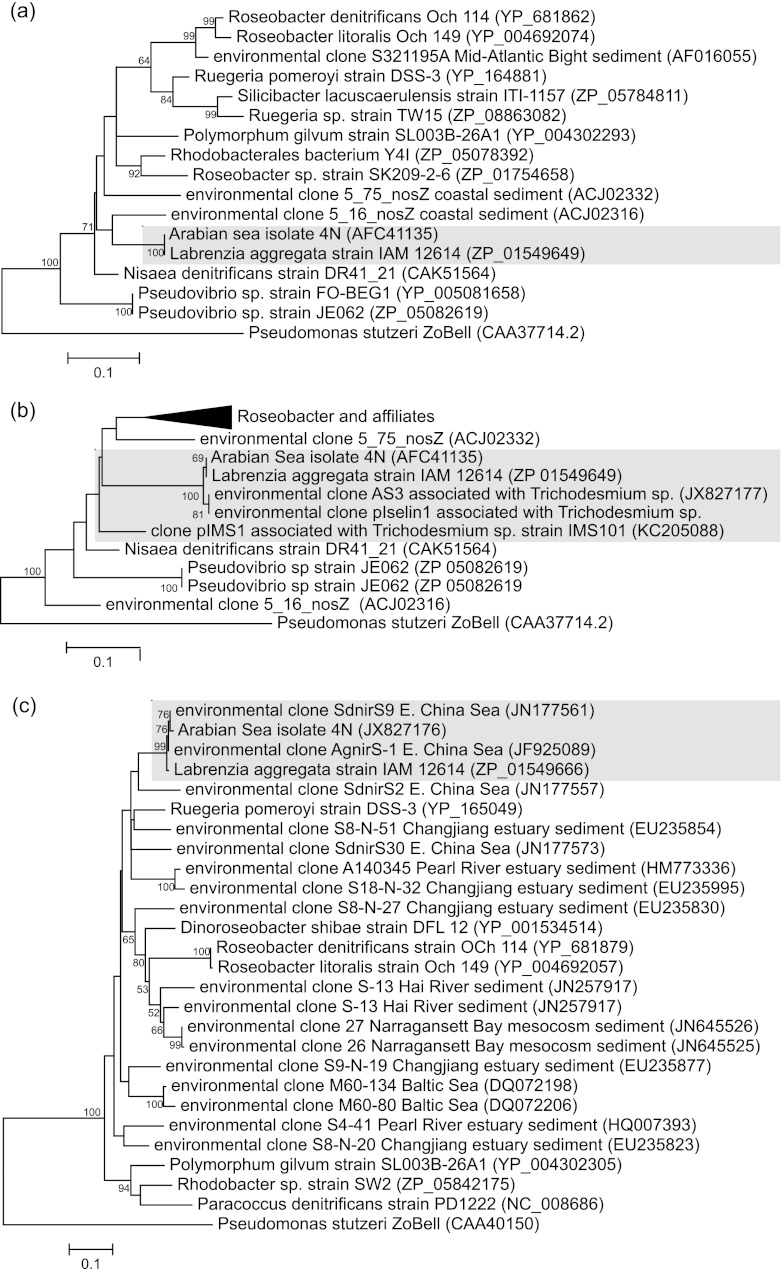
Fig 2.
Phylograms for nosZ (a and b) and nirS (c) based on partial peptide sequences (188, 83, and 283 residues, respectively) from the Arabian Sea isolate 4N and closely related alphaproteobacterial genes. The maximum likelihood trees were inferred from 500 bootstrap replicates in MEGA5 (28) and rooted with the appropriate orthologue from the ZoBell strain of Pseudomonas stutzeri (ATCC 14405). Those partitions that received greater than 50% support in the bootstrap are indicated at their corresponding nodes. The GenBank accession numbers of the sequences are indicated after the sequence names, and the distance scale is shown to the bottom left of each phylogram. The shaded regions highlight the clusters within which genes from the Arabian Sea isolate 4N are found in each phylogram. Note that the highlighted sequence designated pIselin1 in panel b was obtained from Trichodesmium thiebautii DNA collected in 1991 from near-surface waters in the Bahama Islands aboard RV Columbus Iselin during cruise 91-11 (84) and was identical to that of clone AS3 obtained from station 3 in the Arabian Sea (GenBank accession number JX827177). The template used to amplify the nosZ clone pIMS1 from Trichodesmium strain IMS101 was from a DNA sample originating from the laboratory of Jon Zehr (University of California, Santa Cruz) and was supplied by Jon G. Kramer of the Center of Marine Biotechnology, University of Maryland, in late 1997.