Abstract
Saxitoxin and its derivatives are potent neurotoxins produced by several cyanobacteria and dinoflagellate species. SxtA is the initial enzyme in the biosynthesis of saxitoxin. The dinoflagellate full mRNA and partial genomic sequences have previously been characterized, and it appears that sxtA originated in dinoflagellates through a horizontal gene transfer from a bacterium. So far, little is known about the remaining genes involved in this pathway in dinoflagellates. Here we characterize sxtG, an amidinotransferase enzyme gene that putatively encodes the second step in saxitoxin biosynthesis. In this study, the entire sxtG transcripts from Alexandrium fundyense CCMP1719 and Alexandrium minutum CCMP113 were amplified and sequenced. The transcripts contained typical dinoflagellate spliced leader sequences and eukaryotic poly(A) tails. In addition, partial sxtG transcript fragments were amplified from four additional Alexandrium species and Gymnodinium catenatum. The phylogenetic inference of dinoflagellate sxtG, congruent with sxtA, revealed a bacterial origin. However, it is not known if sxtG was acquired independently of sxtA. Amplification and sequencing of the corresponding genomic sxtG region revealed noncanonical introns. These introns show a high interspecies and low intraspecies variance, suggesting multiple independent acquisitions and losses. Unlike sxtA, sxtG was also amplified from Alexandrium species not known to synthesize saxitoxin. However, amplification was not observed for 22 non-saxitoxin-producing dinoflagellate species other than those of the genus Alexandrium or G. catenatum. This result strengthens our hypothesis that saxitoxin synthesis has been secondarily lost in conjunction with sxtA for some descendant species.
INTRODUCTION
Dinoflagellates are a diverse group of unicellular protists that play important ecological roles in marine and freshwater habitats. Approximately 2,000 species of extant dinoflagellates are known to date, most of which are found in marine habitats (1). Dinoflagellates are known for the array of toxic compounds they produce, even though fewer than 100 species have been identified as synthesizing compounds toxic to humans (2). One group of toxins, saxitoxin (STX) and its documented 58 analogues, are potent environmental neurotoxins that can cause the severe human illness paralytic shellfish poisoning (PSP) upon consumption of vector species such as mussels, clams, and oysters (3, 4). Within the dinoflagellates, STX is synthesized by species of the genera Alexandrium, Gymnodinium, and Pyrodinium (see Orr et al., 2011 [5], and references therein). However, the same compounds are also produced by several species of freshwater cyanobacteria (6–8). In both eukaryotes and prokaryotes, STX appears to be synthesized by similar processes; precursor incorporation patterns and stereochemistry are identical in cyanobacteria and dinoflagellates (9–11). The biosynthetic pathway and genes responsible for STX synthesis have been characterized in numerous cyanobacterial species (12–17). The STX biosynthetic pathway in Cylindrospermopsis raciborskii T3, for example, is encoded by more than 35 kb, with 30 catalytic functions being assigned to 26 proteins (14). A complement of 14 genes (sxtA-sxtI, sxtP-sxtR, sxtS, and sxtU) is common between the sxt clusters of five cyanobacterial STX-producing strains (17). These have been subsequently defined as “core” genes (4, 17). Eight of these genes (sxtA, sxtB, sxtD, sxtG, sxtH or sxtT, sxtI, sxtS, and sxtU) seem to be directly implicated in STX synthesis (14). The majority of these have seemingly originated in cyanobacteria via horizontal gene transfers (HGTs) from other bacteria (13).
In dinoflagellates, expressed sequence tags (ESTs) homologous to several cyanobacterial sxt genes have recently been identified in Gymnodinium catenatum and multiple species within the genus Alexandrium (18). In addition, sxtA, the unique starting gene of STX synthesis, has been characterized in detail for Alexandrium fundyense CCMP1719 and Alexandrium minutum CCMP113 (18). The dinoflagellate sxtA gene has a typical dinoflagellate organization (18), it is present in multiple copies in the nuclear genome, its mRNA transcripts are monocistronic as opposed to the cyanobacterial polycistronic sxt transcripts, and it has spliced leader sequences (19) and a eukaryotic polyadenylated tail. Additionally, the GC content of the Alexandrium sxtA transcripts was higher than that of the cyanobacterial sxtA homolog and thus more typical of Alexandrium transcriptomes. These results clearly showed that dinoflagellates carry STX genes themselves and that STX synthesis in dinoflagellates is not due to cocultured bacteria, as previously suggested (20–22).
From a phylogenetic point of view, it appears that STX production is paraphyletic within the genus Alexandrium and that STX-producing (STX+) and -nonproducing (STX−) strains of the same species coexist (5). The presence of sxtA correlates well with the pattern of STX production, although there may be some apparent exceptions (18). This has been attributed to the detection limits of nongenetically based methods (5, 18, 23, 24). Based on the sxtA data, the origin of the STX gene cluster within the dinoflagellates may have occurred through a horizontal gene transfer event between an ancestral STX+ bacterium and the common ancestor of Alexandrium and Pyrodinium (18). To date, this hypothesis is based on sxtA, a single gene in a pathway that may consist of up to 26 proteins (14). In cyanobacteria, the product of sxtA is the substrate for the amidinotransferase SxtG, which is proposed to incorporate an amidino group from a second arginine molecule into the STX intermediate (14). It was the aim of this study to characterize the second core gene of the STX pathway in dinoflagellates, sxtG, and to determine if its structure, phylogeny, and putative evolution correspond to those of sxtA.
MATERIALS AND METHODS
Culturing.
The dinoflagellate strains used in this study (Table 1) were grown in L1 media (25) or GSe media (26) at 16 to 25°C. In addition, Polarella glacialis CCMP2088 was grown at 5°C. All strains were grown with a 12/12-h light/dark photoperiod and a photon irradiance of ∼100 mmol photons m−2 s−1. Strains were not maintained axenically. The identity of each strain was confirmed by amplifying the 18S ribosomal DNA (rDNA) gene using the primer pair NSF83-1528R or 18sF8-ITSR01 (5, 27, 28).
Table 1.
List of dinoflagellate species/strains used in this study and their characteristics regarding production of STX and amplification of sxtA1, sxtA4, and sxtGa
| Order and species | Strain | Production of STX (reference or source) | Amplification of: |
sxtG intron length (bp) | |
|---|---|---|---|---|---|
| sxtA1 and sxtA4 (reference[s] or source) | sxtG (this study) | ||||
| Gonyaulacales | |||||
| A. affine | CCMP112 | ND (5) | ND (18) | + | 260 |
| A. andersoni | CCMP2222 | ND (5) | ND (18) | + | |
| A. insuetum | CCMP2082 | ND (5) | ND (this study) | + | No intron |
| A. minutum | CCMP113 | + (5) | + (18) | + | 740 |
| A. minutum | CS320/01 | + (5) | + (18) | + | 740 |
| A. minutum | CS324/16 | + (5) | + (18) | + | 750 |
| A. tamarense complex | |||||
| A. fundyense (group 1) | CCMP1719 | + (5) | + (18) | + | No intron |
| A. tamarense (group 3) | CCMP1771 | ND (5) | + (18) | + | No intron |
| A. catenella (group 4) | CCMP1493 | + (5) | + (18) | + | 487 |
| A. catenella (group 4) | ACCC01 | + (24) | + (18) | + | 507 |
| A. tamarense (group 5) | ATEB01 | + (23) | + (18, 23) | + | 407 |
| Ceratium longipes | CCMP1770 | ND (50) | ND | ||
| Coolia monotis | CAWD98 | ND (50) | ND | ||
| Gambierdiscus australes | CAWD149 | ND (50) | ND | ||
| Lingulodinium polyedrum | CCMP1931 | ND (50) | ND | ||
| Protoceratium reticulatum | CAWD99 | ND (50) | ND | ||
| Pyrocystis noctiluca | CCMP732 | ND (50) | ND | ||
| Thecadinium kofoidii | SCCAP K-1504 | ND (50) | ND | ||
| Gymnodiniales | |||||
| Amphidinium carteri | UIO081 | ND (50) | ND | ||
| Amphidinium massartii | CS-259 | ND (18) | ND | ||
| Amphidinium mootonorum | CAWD161 | ND (50) | ND | ||
| Gymnodinium aureolum | SCCAP K-1561 | ND (50) | ND | ||
| Gymnodinium catenatum | CCMP1937 | + (52) | + (18) | + | |
| Karlodinium veneficum | RCC2539 | ND (50) | ND | ||
| Lepidodinium chlorophorum | RCC2537 | ND (50) | ND | ||
| Peridiniales | |||||
| Adenoides eludens | CCMP1891 | ND (50) | ND | ||
| Azadinium spinosum | RCC2538 | ND (50) | ND | ||
| Heterocapsa triquetra | RCC2540 | ND (50) | ND | ||
| Pentapharsodinium dalei | SCCAP K-1100 | ND (50) | ND | ||
| Scrippsiella trochoideae | BS-46 | ND (50) | ND | ||
| Prorocentrales | |||||
| Prorocentrum lima | CS-869 | ND (18) | ND | ||
| Prorocentrum micans | UIO292 | ND (50) | ND | ||
| Prorocentrum minimum | UIO085 | ND (50) | ND | ||
| Suessiales | |||||
| Polarella glacialis | CCMP2088 | ND (50) | ND | ||
ND, not detected; +, amplified sequence. The group-naming system of Lilly et al. (2007) is used for A. tamarense complex strains (40).
Nucleic acid isolation and cDNA synthesis.
Genomic DNA (gDNA) and total RNA were isolated from 20-ml cultures in the exponential growth phase, centrifuged for 2 min at 12,000 × g, washed with phosphate-buffered saline (PBS) and bead beaten on dry ice with the FastPrep-24 from Medinor (20 s at speed 4) using 1.4-mm beads (Medinor). The Invitrogen ChargeSwitch gDNA plant kit (Invitrogen) or the Invitrogen ChargeSwitch TotalRNA cell kit (Invitrogen) was utilized in accordance with the supplied protocol. Total RNA from Gymnodinium catenatum (CCMP1937) was kindly donated by Johannes Hagström, Linnaeus University, Kalmar, Sweden. First-strand cDNA was synthesized with the Invitrogen 3′ rapid amplification of cDNA ends (RACE) system (Invitrogen) by following the high-GC protocol and utilizing the adapter primer (AP). DNA, RNA, and cDNA quality was checked with a NanoDrop spectrophotometer (ThermoScientific).
Identification of sxtG homologs and primer design.
Putative sxtG homologs were identified from two previously published Alexandrium 454 high-throughput sequenced cDNA libraries (A. fundyense CCMP1719 and A. minutum CCMP113 [18]) deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the respective accession numbers SRX040427 and SRX040428 (18). Nine A. fundyense CCMP1719 and seven A. minutum CCMP113 putative sxtG contigs were identified as previously described by Stüken et al. (2011) (18). The contigs were blasted (E value, <10) against the nonredundant EST and SRA databases at NCBI (as of January 2012). Orthologous Alexandrium EST (accession numbers EX463008, CK786100, and CK782453) and SRA (accession number SRX111568) sequences were identified and aligned with all sxtG sequences before designing primers. The primers used in this study have been designed using Primaclade (29). Melting temperatures (Tm) were calculated using OligoCalc (30).
PCR, sequencing, and assembly.
PCRs were performed with HotStarTaq Plus polymerase (Qiagen) or BD Advantage 2 Polymerase Mix (BD Biosciences) in the presence of 3% dimethyl sulfoxide (DMSO), 0.2 mM deoxynucleoside triphosphates (dNTPs), and 0.5 mM (each) forward and reverse primers (Table 2) in an MJ Research PTC-200 Thermo Cycler (MJ Research). sxtG PCRs were run under the following cycling conditions: 1 cycle of 5 min at 95°C; 35 cycles of 30 s at 94°C, 30 s at variable temperatures (Table 2), and 1 to 2 min at 72°C; and 1 cycle of 10 min at 72°C. sxtG (genomic and transcript) was amplified with various primer pairs (Table 2).
Table 2.
Primers specifically designed for this study or used in previous studiesa
| Primer name | Primer direction | Primer sequence (5′–3′) | Tm (°C) | Annealing site (5′–3′) | Reference or source |
|---|---|---|---|---|---|
| SL | F | DCCGTAGCCATTTTGGCTCAAG | 57.0 | Variable | 19 |
| AUAP | R | GGCCACGCGTCGACTAGTAC | 57.9 | Variable | Invitrogen |
| Nsf83 | F | GAAACTGCGAATGGCTCATT | 49.7 | 82–101 | 27 |
| 1528r | R | TGATCCTTCTGCAGGTTCACCTAC | 57.4 | 1777–1800 | 28 |
| 18SF8 | F | TTGATCCTGCCAGTAGTCATATGCTTG | 58.2 | 8–34 | 5 |
| ITSR01 | R | CCTTGTTACGACTTCTCCTTCCTC | 57.4 | 1748–1771 | 5 |
| 5.8S-b5′ed | F | GATGAAGAATGCAGCAAMATG | 50.5 | 2025–2045 | 39 |
| 5.8S-b3 | R | CAAGCAHACCTTCAAGMATATCC | 55.3 | 2112–2134 | 39 |
| M13F | F | GTAAAACGACGGCCAG | 45.9 | Invitrogen | |
| M13R | R | CAGGAAACAGCTATGA | 40.8 | Invitrogen | |
| sxt001 | F | TGCAGCGMTGCTACTCCTACTAC | 57.1 | 904–926 | 18 |
| sxt002 | R | GGTCGTGGTCYAGGAAGGAG | 55.9 | 1429–1449 | 18 |
| sxt007 | F | ATGCTCAACATGGGAGTCATCC | 54.8 | 3174–3195 | 18 |
| sxt008 | R | GGGTCCAGTAGATGTTGACGATG | 57.1 | 3865–3888 | 18 |
| sxtG71F | F | AGGACATGGACGAKAATAGCTG | 54.8 | 71–92 | This study |
| sxtG127F | F | TCCGGCGACTACGAGTTC | 52.6 | 127–144 | This study |
| sxtG206F | F | GGGCCGTGAAGGATTACCTGA | 56.3 | 206–226 | This study |
| sxtG206R | R | TCAGGTAATCCTTCACGGCCC | 56.3 | 206–226 | This study |
| sxtG221R | R | GTTTATGCCGTCGCGCTTCAGGT | 58.8 | 221–243 | This study |
| sxtG663F | F | CATGGAGTCGATGGTGAGCAAC | 56.7 | 663–684 | This study |
| sxtG663R | R | GTTGCTCACCATCGACTCCATG | 56.7 | 663–684 | This study |
| sxtG802F | F | CTGGACTCGMACACGATAATGA | 54.8 | 802–823 | This study |
| sxtG1008F | F | CGAGTCCTACGGCTACAAGC | 55.9 | 1008–1027 | This study |
| sxtG1009R | R | ATCGGCTTGTAGCCGTAGGACTC | 58.8 | 1009–1031 | This study |
| sxtG1042R | R | CGCAGAAGTTCATGTYGCATAT | 53.0 | 1042–1063 | This study |
| sxtGq559F | F | GACGGGAACGGCTACAA | 49.5 | 559–575 | This study |
| sxtGq605R | R | GCTCGAAGATCGGGTCCT | 52.6 | 605–622 | This study |
Melting temperature (Tm) calculated using OligoCalc (30). F, forward; R, reverse. Annealing sites are based on the mRNA sequence of A. fundyense and can vary slightly between species. sxtG intron presence is not considered. The primer pairs and PCR annealing temperature used were as follows: 60°C for sxtG127F/sxtG1009R, sxtG206F/sxtG1009R, sxtG127F/sxtG663R, sxtG206F/sxtG663R, and sxtG663F/sxtG1009R, and 56°C for sxtG71F/sxtG663R and sxtG663F/sxtG1042R.
The entire transcript was amplified for A. fundyense CCMP1719 and A. minutum CCMP113. The 5′ end of the transcript, including the dinoflagellate spliced leader sequence, was amplified using the primers dinoSL (19) and sxtG663R. The 3′ end, including the eukaryotic poly(A) tail, was amplified using the sxtG206F and the AUAP adaptor primers (Invitrogen). The 5′ and 3′ end products were cloned with TOPO TA (Invitrogen) before being sequenced with the M13 forward and reverse primers. Additionally, genomic amplicons of Alexandrium affine CCMP112 and A. fundyense CCMP1719 were cloned, as direct sequencing indicated the presence of divergent sxtG copies within these two strains. Amplification of sxtG was checked for all non-PSP-producing dinoflagellates listed in Table 1 (genomic and transcript) using the primer combinations as previously described (Table 2). Positive (STX+ strain CCMP113 or CCMP1719) and nontemplate controls were run in all cases. PCR products were gel excised using the Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced directly using an ABI3730 DNA analyzer (Applied Biosystems). Respective forward and reverse primers as well as several internal primers were utilized for sequencing (Table 2). Sequences were quality checked and assembled using the Phred/Phrap/Consed (31) package under the default settings. Further manual editing was performed in MacClade v4.07 (32).
Additional sxtA PCRs were performed on Alexandrium insuetum CCMP2082 as described in reference 18. For the sxtA1 fragment, primers sxt001 and sxt002 (∼550 bp) were used, and for the sxtA4 fragment, sxt007 and sxt008 (∼750 bp; Table 2) were used.
sxtG analyses and phylogenetic inference.
Dinoflagellate sxtG transcript structure was determined by aligning the translated mRNA sequence with that of sxtG from the cyanobacterium Anabaena circinalis AWQC131C (DQ787201). In addition, conserved domain searches (http://pfam.sanger.ac.uk/search/sequence [33]) were carried out. The open reading frame (ORF) was predicted using a tool available online (http://proteomics.ysu.edu/tools/OrfPredictor.html [34]). Catalytic and substrate-binding residues of sxtG from cyanobacteria have been previously determined (14). An amino acid alignment of the dinoflagellate sxtG coding sequences, orthologous cyanobacterial sxtG sequences, and a selection of closely related NCBInr BLASTP hits (September 2012) was constructed using MAFFTv6 L-INS-I model under the default settings (35). The resulting alignment was checked manually, and poorly aligned positions were excluded using MacClade v4.07 (32). ProtTest v2.4 (36) determined LG as the optimal evolutionary model. Maximum likelihood (ML) analyses were performed with the RAxML-VI-HPC v7.2.6, PROTCATLG model with 25 rate categories (37). The most likely topology was established from 100 separate searches, and bootstrap analyses were performed with 500 pseudoreplicates.
Dinoflagellate genomic sxtG structure was determined by alignment against the corresponding transcript sequence using MacClade v4.07 (32). The comparison of genomic DNA to cDNA identified a splice site at nucleotide (nt) 475 (amino acid [aa] 159) for numerous species. Intron sequence identities and percentages of sequence identity were compared between and within species at the nucleotide level by pairwise alignment using CLC Main Workbench (CLC bio, Aarhus, Denmark) (Table 3). All model estimation and phylogenetic analyses were done on the freely available Bioportal (38) at the University of Oslo (http://www.bioportal.uio.no/).
Table 3.
Comparison of sxtG intron sequence identity and percent identity between species and/or strains
| Species and strain (intron length in bp) | Strain ID | % identity or no. of identities between intron sequencesa |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| A. minutum CCMP113 (740) | 1 | 99.19 | 98.53 | 27.79 | 38.59 | 42.95 | 43.37 | 39.06 | |
| A. minutum CS320/01 (740) | 2 | 734 | 98.00 | 27.79 | 38.72 | 43.21 | 43.62 | 39.19 | |
| A. minutum CS324/16 (750) | 3 | 739 | 735 | 27.43 | 38.10 | 42.77 | 43.18 | 38.56 | |
| A. affine CCMP112 (260) | 4 | 209 | 209 | 209 | 41.87 | 33.33 | 33.21 | 39.33 | |
| A. fundyense CCMP1719 (368) | 5 | 296 | 297 | 296 | 175 | 58.16 | 60.38 | 72.30 | |
| A. catenella CCMP1493 (487) | 6 | 335 | 337 | 337 | 177 | 303 | 93.89 | 75.15 | |
| A. catenella ACCC01 (507) | 7 | 340 | 342 | 342 | 181 | 317 | 476 | 77.71 | |
| A. tamarense ATEB01 (407) | 8 | 300 | 301 | 300 | 175 | 308 | 378 | 394 | |
The upper pairwise comparison values represent percent identity between intron sequences. Unshaded cells indicate >90% identity; shaded cells indicate <80% identity. The lower pairwise comparison values represent the number of identities between intron sequences.
Relative copy number determination.
Quantitative PCR (qPCR) primers were designed based on the sxtG gDNA sequence of A. fundyense CCMP1719 (JX995118) using the Universal Probe Library Design Software from Roche, version 2.45, using default parameters. Suggested primers were in silico tested against an alignment of all available Alexandrium sxtG mRNA and gDNA sequences. The chosen primers annealed to a conserved region downstream of the sxtG intron. The assay was optimized on strains CCMP1493 (A. catenella) and CCMP113 (A. minutum), using different annealing temperatures (55, 60, and 64°C) and primer concentrations (125 nM and 250 nM), assessing crossing point (CP) values and melting curves, and running the PCR fragments on agarose gels. The best results were obtained with the primer pair sxtGq559F/sxtGq605R (Table 2) with annealing temperatures of 60 or 64°C and 125 nM primer concentrations. PCR inhibitions were observed when there was >10 ng input DNA in 20-μl reaction volumes (data not shown). DNA was quantified with a Qubit Fluorometer using the double-stranded DNA (dsDNA) HS assay (Invitrogen). All PCRs were run on a Roche LightCycler480 system in white 96-well plates with Roche SYBR green I Master chemistry. For sxtG qPCR, 20-μl reaction mixtures contained 5 μl 2× SYBR green master mix, 125 nM each primer, and 1 to 10 ng gDNA. They were run with the following protocol: hot start, 1 cycle of 95°C for 10 min; amplification, 45 cycles of 94°C for 10 s, 64°C for 20 s, and 72°C for 10 s, with single acquisition; followed by the melting curve program, 1 cycle of 95°C for 5 s and 65°C for 1 min, with up to 97°C continuous measurements; and finally, cooling, 1 cycle of 40°C for 10 s. qPCRs (5.8S) were run according to Galluzzi et al. (2004) (39) with the slightly modified forward primer 5.8S-b5′ed and the 5.8S-b3′ reverse primer (Table 2). Reaction mixtures (20 μl) contained 5 μl 2× SYBR green master mix, 150 nM (each) primer, and 4 ng gDNA and were run on the following program: hot start, 1 cycle of 95°C for 10 min; amplification, 45 cycles of 95°C for 15 s and 60°C for 45 s, with single acquisition; followed by the same melting and cooling program as for sxtG. As a standard, DNA from strain CCMP1493 (Alexandrium catenella) was diluted to 10, 5, 2.5, 1.25, and 0.63 ng/DNA per reaction. The efficiency of both qPCRs was 90 to 95% using the 2nd derivative method implemented in the LightCycler 480 software release 1.5.0.
Nucleotide sequence accession numbers.
All sxtG sequences generated in this study have been deposited in GenBank under the accession numbers JX995111 to JX995130 (see Table S1 in the supplemental material).
RESULTS
Sequence amplification and assembly.
The sxtG primers designed in this study (Table 2) successfully amplified sxtG fragments in eight Alexandrium species (with A. tamarense group 3 and group 5 being considered separate species; see Orr et al., 2011 [5]), 11 Alexandrium strains (both STX+ and STX−), and Gymnodinium catenatum (Table 1). The assembly of these fragments resulted in sxtG sequences ranging from 375 bp in G. catenatum CCMP1937 to 934 bp in A. affine CCMP112, with a complete ORF for A. fundyense CCMP1719 and A. minutum CCMP113 (see Table S1 in the supplemental material for a complete listing). No sxtG PCR products were amplified for 22 STX− dinoflagellates in five orders (Table 1), while their 18S rDNA sequences were readily amplified from the same DNA. In addition, no eukaryotic sxtG sequence was identified by BLAST against the NCBI nonredundant, EST, and SRA databases in any species other than those of the genus Alexandrium (accession numbers CK786100, CK782453, EX463008, JV310276, and SRX111568).
SxtA PCR.
sxtA1 and sxtA4 were not detected in A. insuetum CCMP2082. The 18S rDNA control was amplified, as were the sxtA1 and sxtA4 positive controls.
Identification and determination of sxtG transcript structure.
Assembly of the nine A. fundyense CCMP1719 and seven A. minutum CCMP113 sxtG contigs identified by Stüken et al. (2011) (18) gave a full ORF for both species, although dinoflagellate spliced leader sequences and poly(A) tails were lacking.
The RACE experiments resulted in full-length sxtG transcripts for both A. fundyense CCMP1719 and A. minutum CCMP113. This included a dinoflagellate spliced leader sequence at the 5′ end and a eukaryotic poly(A) tail at the 3′ end. The transcripts were 1,283 bp and 1,276 bp in length, respectively, excluding the poly(A) tail (Fig. 1). Additionally, the A. tamarense SRA contig (SRX111568) contained 9 bp of the 22-bp dinoSL sequence. Conserved domain searches determined that these putative sxtG sequences encode an amidinotransferase enzyme, as expected. The ORF was 375 amino acids (1,125 bp) long, with a GC content of 64.5% in A. fundyense and 63.4% in A. minutum. In addition, one A. fundyense (JX995134) and three A. minutum contigs (JX995131 to JX995133) identified by Stüken et al. (2011) (18) were predicted as encoding a homologous amidinotransferase.
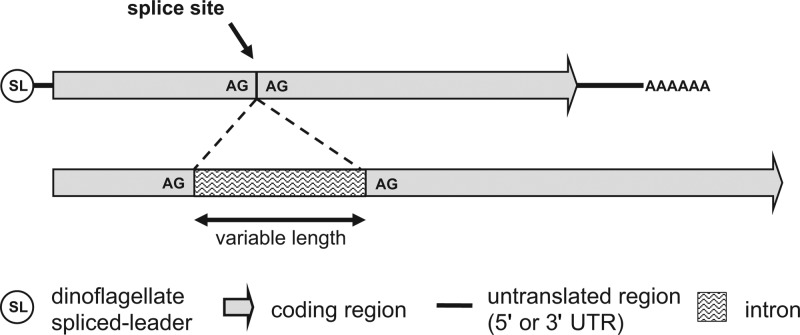
Fig 1.
Structure of sxtG in dinoflagellates. (Top) Transcript structure of sxtG; (bottom) genomic structure of sxtG.
sxtG genomic structure.
Comparisons of transspliced leader amplified cDNA and genomic amplicons did not show any signs of RNA editing. A single splice site at nt 475 (aa 159) with the dinucleotide AG 5′ and 3′ end boundaries was present in numerous species (Table 1; Fig. 1). The intron sequence has several short palindromic repeats and stop codons in all forward reading frames (standard codon table). The removal of the intron resulted in the restoration of the ORF. Intron length varied from 260 bp in A. affine CCMP112 to 750 bp in A. minutum CS 324/16 (Table 1). Intron intraspecies pairwise percent identity was >90%, while interspecies percent identity was <80% (Table 3). No variation in intron sequence was observed within a strain, despite multiple amplifications and sequencing. We were unable to determine intron presence for A. andersoni CCMP222. Likewise, only cDNA for G. catenatum CCMP1937 was amplified in this study.
The cloning of sxtG for A. affine CCMP112 and A. fundyense CCMP1719 revealed a second gene, most likely a pseudogene, not present in the cDNA. For A. fundyense CCMP1719 (JX995130), the pseudogene sequence had a 369-bp intron at nt 475 that diverged from the functional copy at nt 658. The divergent sequence had stop codons in the reading frame (standard code). In the case of A. affine CCMP112 (JX995129), the pseudogene sequence had a 260-bp intron and, in congruence with that of A. fundyense CCMP179, diverged at nt 658. However, no stop codons were observed (standard code).
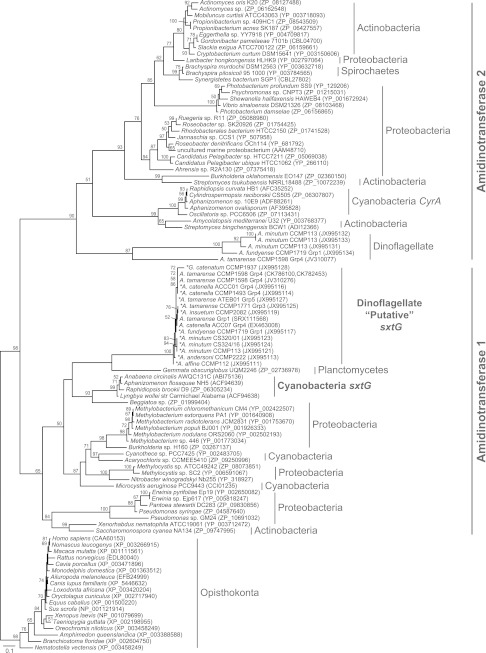
The phylogeny of sxtG.
For interpretation of the phylogenetic inference (Fig. 2), statistical support is defined as follows: full, 100 bootstrap value (BP); high, >90 BP; moderate, >65 BP; and low, >50 BP. The phylogenetic inference of sxtG-encoded amino acid sequences shows that dinoflagellate sxtG sequences form a fully supported clade (Fig. 2). The branching pattern within the dinoflagellate clade is unclear, with minimal support between species and strains. The A. minutum strains form a highly supported monophyletic clade (94 BP), while species and strains within the A. tamarense complex place paraphyletic G. catenatum branches with moderate support (86 BP) within a clade formed by group 4 A. tamarense complex strains, as defined by Lilly et al. (2007) (40). The STX− species A. affine was basal, with A. andersoni next to diverge; however, this placement was not supported. The STX− A. insuetum formed a moderately supported (76 BP) clade with a group 3 A. tamarense. The planctomycete Gemmata obscuriglobus is the sister group to the dinoflagellate sxtG clade; however, this grouping lacked support. The proteobacterium Beggiatoa further excludes the dinoflagellate sequences from a sister relationship with the highly supported cyanobacterial sxtG clade (99 BP). The four previous clades form a low-support (50 BP) group with a cluster of cyanobacterium and proteobacterium species. This is further included in a low-support (65 BP) monophyly with an actinobacterium and proteobacterium clade, constituting the cluster defined as amidinotransferase 1. The amidinotransferase 1 clade is excluded from opisthokonta and the amidinotransferase 2 clades with high support, 98 BP and 90 BP, respectively. The amidinotransferase 2 clade harbors the additional homologous dinoflagellate amidinotransferase sequences identified by Stüken et al. (2011) (18). This fully supported clade forms a moderately supported (87 BP) grouping with an additional A. tamarense amidinotransferase (JV310077). The sister grouping to this dinoflagellate clade consists of actinobacteria and the cyanobacterial aoaA and cyrA genes, involved in the synthesis of cylindrospermopsin (41).
Fig 2.
sxtG phylogenetic tree. Single-gene phylogeny inferred from 363 amino acid characters. The tree is reconstructed with maximum likelihood inference (RAxML). Numbers on the internal nodes represent bootstrap (BP) values of >50%. *, taxon sequences generated from this study. Group 1 to group 5 (Grp 1 to Grp 5) denotations are from the A. tamarense complex group naming system (40).
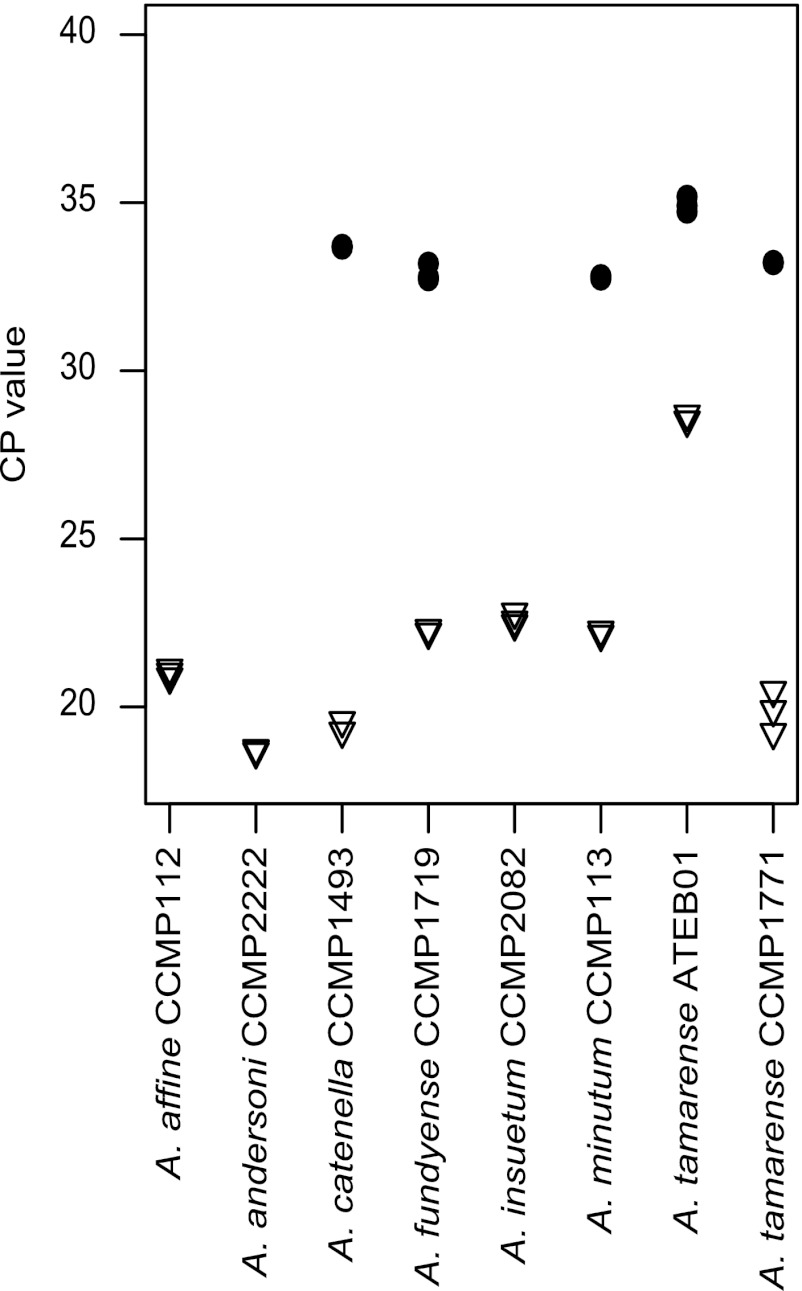
Relative copy number determination.
All strains tested were positive in the 5.8S qPCR (Fig. 3). However, only the strains of the STX+ species A. tamarense, A. fundyense, A. catenella, and A. minutum had positive amplification in the sxtG qPCR (Fig. 3). The remaining STX− strains (A. affine, A. andersoni, and A. insuetum) had repeatedly unspecific melting curves. sxtG is present in considerably fewer copies in the genome than rDNA, though variance of copy number between the strains was relatively constant per ng DNA (Fig. 3).
Fig 3.
Relative copy number results, showing crossing point (CP) values obtained with 4 ng of input gDNA. Triangles represent 5.8S rDNA, and circles represent sxtG.
DISCUSSION
We have previously identified putative dinoflagellate ESTs homologous to cyanobacterial sxtG genes (14, 18). Building on this work, we characterized the entire mRNA transcript sequences as well as partial genomic gene fragments of sxtG in dinoflagellates in the present work. Our results show that the putative sxtG ESTs originally identified by Stüken et al. (2011) (18) have a typical dinoflagellate structure and are the most likely candidates to be the dinoflagellate homologs of the cyanobacterial sxtG genes.
The transcriptomic and genomic structure of sxtG.
We have amplified the entire sxtG mRNA transcripts from A. fundyense CCMP1719 and A. minutum CCMP113 and partial transcripts from five strains. In contrast to the bacterial transcripts, the dinoflagellate sxtG transcripts were monocistronic and possessed poly(A) tails and dinoflagellate spliced leader sequences (Fig. 1). The result demonstrates that both the first gene and the second gene of the STX pathway are borne in the nuclear genome of dinoflagellates (18). In congruence with sxtA, the GC content of sxtG was ∼20% higher in dinoflagellates (with a GC of ∼64%) than for the STX+ cyanobacterium species, suggesting that the gene has been modified extensively since its introduction into the dinoflagellates (18).
The genomic sequences of sxtG contain noncanonical introns that lack a GT/AG splicing site and vary in length and sequence between various Alexandrium species (Table 1; Fig. 1). The introns show high interspecies and low intraspecies variability, with no observed variation within a strain (Table 3). This may prove useful when trying to identify strains within species or to differentiate between species of Alexandrium. For example, the low percent identity between the A. tamarense complex group 4 strains (ACCC01 and CCMP1493) and group 5 strain (ATEB01) confirms that these two groups are different enough to be considered separate species (5, 40). The confirmed presence of introns in sxt genes may also explain the amplification problems of the full-length genomic sxtA sequence (A. Stüken, unpublished data).
It is thought that introns are rare in dinoflagellate genomes, as a gene should be purged of introns with every passage through the DNA-RNA cycle (42). However, to date, when introns have been found, most lack a GT/AG splice site and are present only in a selection of species (43–47). Considering the variation in sequence length and identity (Table 3) and that the intron is not present for all species, multiple independent losses and acquisitions of the sxtG intron are likely. This “birth and death” pattern of gene evolution has also been observed for actin genes in Dinophysis species (48) and is supported by the presence of nonfunctional sxtG copies at least in the case of A. fundyense CCMP1719.
Occurrence and distribution of sxtG.
Unlike with sxtA, the presence of sxtG is not specific to species known to produce STX. The sxtG amidinotransferase was present and transcribed in all tested Alexandrium species, including those for which sxtA and STX synthesis have not been detected (5, 18). However, some inconsistencies to this pattern were observed. First, it is uncertain if the divergent sxtG genomic copy amplified from A. affine CCMP112 is functional, as no additional copy was detected. No stop codons were observed in the sequence. We cannot exclude the possibility that the gene would be extensively modified posttranscriptionally, for example, by mRNA editing, though this may seem unlikely, as all other strains appeared free from editing.
Further, while qPCR results showed the sxtG copy number to be relatively constant between the known STX+ species, no or unspecific amplification was observed for the STX− species (Fig. 3). It is unclear why qPCR and conventional PCR results for the STX− species are discrepant. It is possible that low genomic sxtG copy numbers, large genome sizes, or a combination of these factors may account for this. For example, the predicted DNA content per cell of A. affine CCMP112 (A. Stüken and Rosa I. Figueroa, unpublished data) is higher than that of A. tamarense CCMP1598, which is estimated at 103.5 pg DNA cell−1 (49). A low-copy-number gene in such a large genome may be difficult to detect. However, the results for A. andersoni CCMP2222, with 21.8 pg DNA cell−1, and A. insuetum CCMP2082, with 30.8 pg DNA cell−1 (49), are not in line with such an explanation.
Consistent with sxtA distribution (18, 50), we did not detect sxtG in non-PSP dinoflagellate genera other than the Alexandrium genus and Gymnodinium catenatum (Table 1). The presence of a divergent, nonhomologous sxtG sequence cannot be discounted for non-PSP dinoflagellate genera. However, searches of multiple GenBank databases gave sxtG hits to Alexandrium only, suggesting that it was not present in the other organisms. The fact that STX− Alexandrium spp. carry and transcribe sxtG may suggest that this gene is also involved in other biochemical pathways or that the capacity for STX synthesis has been comparatively recently lost in these species.
The analysis of sxtG in Pyrodinium bahamense would be desirable, as it produces saxitoxin. However, cultures of this species are currently not publicly available. The presence of both sxtA and sxtG in this species has recently been reported, though the sequences are yet to be made available in GenBank (51). The P. bahamense sxtG sequences presented by Hackett et al. (2013), however, group phylogenetically with the A. tamarense group 4 strain CCMP1598, contig 87049 (51), JV310276 in Fig. 2, indicating that the amino acid sequences are similar.
The phylogeny of sxtG.
The phylogenetic inference of sxtG (Fig. 2) shows that dinoflagellate sxtG sequences form a fully supported clade. The branching pattern within the clade is unclear, a result of high sequence conservation of the coding sequence between species. Even at the nucleotide level (results not shown), sequences are highly conserved, limiting sxtG as a phylogenetic marker. Thus, it was not possible to determine if the evolution of sxtG mirrors that of sxtA or even the Alexandrium genus (5, 18). As the entire ORF was not amplified from all species, lower conservation cannot be completely rejected, with putative degenerate binding sites being an explanation.
The planctomycete Gemmata obscuriglobus was found to be the sister group to the dinoflagellate sxtG clade. However, it is difficult to make conclusions regarding the relationships, as the clades were poorly supported. It appears that this dinoflagellate amidinotransferase protein, congruent with sxtA, was not acquired directly from cyanobacteria (18). It is difficult to conclude if sxtG was attained independently of sxtA, with the inferred phylogenies lacking resolution (18). Considering the inference in Fig. 2, this common ancestor may be, as previously suggested for the cyanobacterial homolog, a proteobacterium (13). A second dinoflagellate amidinotransferase that groups more distantly to sxtG with homologous actinobacterial and cyanobacterial cylindrospermopsin aoaA and cyrA sequences was additionally identified. This may suggest that multiple amidinotransferases have been acquired by HGT in parallel or separate events. It is not clear what pathway, if any, these genes are involved in. As dinoflagellates have been found to synthesize a multitude of different compounds, it is possible that they are involved in the synthesis of an unrelated compound.
The origin of STX in dinoflagellates.
The origin of the biosynthetic pathway and genes responsible for STX synthesis in the dinoflagellates has been proposed through an HGT event with an ancestral STX+ bacterium (18). The hypothesis, at present based on the starting gene of STX synthesis, sxtA, proposes that the HGT probably occurred before Alexandrium and Pyrodinium diverged within the order Gonyaulacales. Thus, STX synthesis may have been secondarily lost for some descendant species. Gymnodinium catenatum, order Gymnodiniales, which has an sxtA sequence that is conserved to and branches within the Alexandrium genus, probably independently acquired STX from a later dinoflagellate-dinoflagellate transfer (18). This is further supported by recent improvements in the resolution of dinoflagellate order relationships and the nondetection of sxtA for multiple non-PSP-producing dinoflagellate species (50).
The results presented here add support to this hypothesis. However, sxtG, in contrast to sxtA, is not exclusive to STX+ species but is present in all Alexandrium species. As far as we can deduce from our results, sxtG is absent from non-PSP-producing dinoflagellate genera and may have so few genomic copies in STX− Alexandrium species that it is difficult to detect via qPCR. This would suggest that some Alexandrium species might have had the capacity to synthesize STX until the secondary loss of essential genes, possibly in unison with sxtA. Considering the rDNA phylogeny of Alexandrium, it appears that sxtA has been lost within the genus on multiple occasions independently, rather than from a single event (5). If an HGT occurred prior to the split of Alexandrium and Pyrodinium, we may expect other possible descendant genera to also produce sxtG. Considering the broadly sampled dinoflagellate phylogenies presented by Orr et al. (2012), Ceratium, Gamberdiscus, and Pyrocystis may all have had this capacity (50). The apparent lack of sxtG for these genera may indicate an ancient HGT event, with non-PSP-producing Alexandrium having only recently lost the ability to synthesize STX with the loss of sxtA. Multiple separate events cannot be discounted, though this seems unlikely.
Congruent with phylogenies based on sxtA1 and sxtA4 (18), the G. catenatum sxtG sequence branches within the Alexandrium genus (Fig. 2). This further strengthens a secondary dinoflagellate-dinoflagellate transfer theory for the origin of STX in this species (18), although additional sxtG sequences for G. catenatum are needed to assess this fully. In addition, the lack of both a homologous sxtA gene and a homologous sxtG gene for Lepidodinium chlorophorum, the possible sister taxon to G. catenatum, may add more support to this scenario (50).
Despite these advances, it is difficult to conclude fully on the evolutionary origin of the STX pathway in dinoflagellates at present. The phylogenetic relationships within both the Gonyaulacales order and the Alexandrium genus need to be more robustly resolved (5, 50). In addition, previously identified sxt homologs need to be characterized in detail to resolve the origin of STX in dinoflagellates (18).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Shuhei Ota and the Section of Marine Biology, Department of Biosciences, UiO, for access to cultures. Likewise, Gustaaf Hallegraeff and Lesley Rhodes are thanked. We are indebted to Johannes Hagström for providing us with total RNA from G. catenatum (CCMP1937) and to Rouna Yauwenas for technical help. Two anonymous reviewers are thanked for comments that helped improve this paper.
This study was financially supported by NFR grant 186292/V40 to K.S.J. and by a Ph.D. grant to R.J.S.O. from the Molecular Life Science program (MLS), University of Oslo.
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03279-12.
REFERENCES
- 1. Taylor FJR. 1987. General group characteristics, special features of interest, short history of dinoflagellate study, p 798 In Taylor FJR. (ed), The biology of dinoflagellates. Botanical monographs, vol 21 Blackwell Scientific Publications, Oxford, United Kingdom [Google Scholar]
- 2. Hallegraeff GM. 1993. A review of harmful algal blooms and their apparent global increase. Phycologia 32:79–99 [Google Scholar]
- 3. Deeds JR, Landsberg JH, Etheridge SM, Pitcher GC, Longan SW. 2008. Non-traditional vectors for paralytic shellfish poisoning. Mar. Drugs 6:308–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wiese M, D'Agostino PM, Mihali TK, Moffitt MC, Neilan BA. 2010. Neurotoxic alkaloids: saxitoxin and its analogs. Mar. Drugs 8:2185–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orr RJS, Stüken A, Rundberget T, Eikrem W, Jakobsen KS. 2011. Improved phylogenetic resolution of toxic and non-toxic Alexandrium strains using a concatenated rDNA approach. Harmful Algae 10:676–688 [Google Scholar]
- 6. Alam M, Ikawa M, Sasner JJ, Sawyer PJ. 1973. Purification of Aphanizomenon flos-aquae toxin and its chemical and physiological properties. Toxicon 11:65–72 [DOI] [PubMed] [Google Scholar]
- 7. Schantz EJ, Lynch JM, Vayvada G, Matsumot K, Rapoport H. 1966. Purification and characterization of poison produced by Gonyaulax catenella in axenic culture. Biochemistry 5:1191–1195 [DOI] [PubMed] [Google Scholar]
- 8. Smith JL, Boyer GL, Zimba PV. 2008. A review of cyanobacterial odorous and bioactive metabolites: impacts and management alternatives in aquaculture. Aquaculture 280:5–20 [Google Scholar]
- 9. Shimizu Y. 1996. Microalgal metabolites: a new perspective. Annu. Rev. Microbiol. 50:431–465 [DOI] [PubMed] [Google Scholar]
- 10. Shimizu Y. 1993. Microalgal metabolites. Chem. Rev. 93:1685–1698 [Google Scholar]
- 11. Kellmann R, Stüken A, Orr RJS, Svendsen HM, Jakobsen KS. 2010. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 8:1011–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mihali T, Kellmann R, Neilan B. 2009. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 10:8 doi:10.1186/1471-2091-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moustafa A, Loram JE, Hackett JD, Anderson DM, Plumley FG, Bhattacharya D. 2009. Origin of saxitoxin biosynthetic genes in cyanobacteria. PLoS One 4:e5758 doi:10.1371/journal.pone.0005758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, Neilan BA. 2008. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 74:4044–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stucken K, John U, Cembella A, Murillo AA, Soto-Liebe K, Fuentes-Valdes JJ, Friedel M, Plominsky AM, Vasquez M, Glockner G. 2010. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS One 5:e9235 doi:10.1371/journal.pone.0009235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mihali TK, Carmichael WW, Neilan BA. 2011. A putative gene cluster from a Lyngbya wollei bloom that encodes paralytic shellfish toxin biosynthesis. PLoS One 6:e14657 doi:10.1371/journal.pone.0014657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murray SA, Mihali TK, Neilan BA. 2011. Extraordinary conservation, gene loss, and positive selection in the evolution of an ancient neurotoxin. Mol. Biol. Evol. 28:1173–1182 [DOI] [PubMed] [Google Scholar]
- 18. Stüken A, Orr RJS, Kellmann R, Murray SA, Neilan BA, Jakobsen KS. 2011. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS One 6:e20096 doi:10.1371/journal.pone.0020096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang H, Hou YB, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin SJ. 2007. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U. S. A. 104:4618–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kodama M, Ogata T, Sakamoto S, Sato S, Honda T, Miwatani T. 1990. Production of paralytic shellfish toxins by a bacterium Moraxella sp. isolated from Protogonyaulax tamarensis. Toxicon 28:707–714 [DOI] [PubMed] [Google Scholar]
- 21. Gallacher S, Flynn KJ, Franco JM, Brueggemann EE, Hines HB. 1997. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp (Dinophyta) in culture. Appl. Environ. Microbiol. 63:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vasquez M, Gruttner C, Gallacher S, Moore ERB. 2001. Detection and characterization of toxigenic bacteria associated with Alexandrium catenella and Aulacomya ater contaminated with PSP. J. Shellfish Res. 20:1245–1249 [Google Scholar]
- 23. Murray SA, Wiese M, Neilan BA, Orr RJS, de Salas M, Brett S, Hallegraeff G. 2012. A reinvestigation of saxitoxin production and sxtA in the ‘non-toxic’ Alexandrium tamarense Group V clade. Harmful Algae 18:96–104 [Google Scholar]
- 24. Murray SA, Wiese M, Stüken A, Brett S, Kellmann R, Hallegraeff G, Neilan BA. 2011. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 77:7050–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guillard RRL, Hargraves PE. 1993. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32:234–236 [Google Scholar]
- 26. Blackburn SI, Bolch CJS, Haskard KA, Hallegraeff GM. 2001. Reproductive compatibility among four global populations of the toxic dinoflagellate Gymnodinium catenatum (Dinophyceae). Phycologia 40:78–87 [Google Scholar]
- 27. Hendriks L, Goris A, Neefs JM, Van de Peer Y, Hennebert G, Dewachter R. 1989. The nucleotide sequence of the small ribosomal-subunit RNA of the yeast Candida albicans and the evolutionary position of the Fungi among the Eukaryotes. Syst. Appl. Microbiol. 12:223–229 [Google Scholar]
- 28. Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499 [DOI] [PubMed] [Google Scholar]
- 29. Gadberry MD, Malcomber ST, Doust AN, Kellogg EA. 2005. Primaclade—a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 21:1263–1264 [DOI] [PubMed] [Google Scholar]
- 30. Kibbe WA. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35:W43–W46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195–202 [DOI] [PubMed] [Google Scholar]
- 32. Maddison WP, Maddison DR. 2000. MacClade, 4th ed Sinauer Associates, Sunderland, MA [Google Scholar]
- 33. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Min XJ, Butler G, Storms R, Tsang A.2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33:W677–W680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katoh K, Toh H. 2008. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform. 9:212 doi:10.1186/1471-2105-9-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 37. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 38. Kumar S, Skjaeveland A, Orr RJS, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. 2009. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinform. 10:357 doi:10.1186/1471-2105-10-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galluzzi L, Penna A, Bertozzini E, Vila M, Garcés E, Magnani M. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lilly EL, Halanych KM, Anderson DM. 2007. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae). J. Phycol. 43:1329–1338 [Google Scholar]
- 41. Muenchhoff J, Siddiqui KS, Poljak A, Raftery MJ, Barrow KD, Neilan BA. 2010. A novel prokaryotic L-arginine:glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 277:3844–3860 [DOI] [PubMed] [Google Scholar]
- 42. Slamovits CH, Keeling PJ. 2008. Widespread recycling of processed cDNAs in dinoflagellates. Curr. Biol. 18:R550–R552 [DOI] [PubMed] [Google Scholar]
- 43. Okamoto OK, Liu LY, Robertson DL, Hastings JW. 2001. Members of a dinoflagellate luciferase gene family differ in synonymous substitution rates. Biochemistry 40:15862–15868 [DOI] [PubMed] [Google Scholar]
- 44. Rowan R, Whitney SM, Fowler A, Yellowlees D. 1996. Rubisco in marine symbiotic dinoflagellates: form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 8:539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H, Lin SJ. 2003. Complex gene structure of the form II RUBISCO in the dinoflagellate Prorocentrum minimum (Dinophyceae). J. Phycol. 39:1160–1171 [Google Scholar]
- 46. Hoppenrath M, Leander BS. 2010. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS One 5:e13220 doi:10.1371/journal.pone.0013220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bachvaroff TR, Place AR. 2008. From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS One 3:e2929 doi:10.1371/journal.pone.0002929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S, Bachvaroff TR, Handy SM, Delwiche CF. 2011. Dynamics of actin evolution in dinoflagellates. Mol. Biol. Evol. 28:1469–1480 [DOI] [PubMed] [Google Scholar]
- 49. LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. 2005. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 41:880–886 [Google Scholar]
- 50. Orr RJS, Murray S, Stüken A, Rhodes L, Jakobsen KS. 2012. When naked became armored: an eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS One 7:e50004 doi:10.1371/journal.pone.0050004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hackett JD, Wisecaver JH, Brosnahan ML, Kulis DM, Anderson DM, Bhattacharya D, Plumley FG, Erdner DL. 2013. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 30:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ordás MC, Fraga S, Franco JM, Ordás A, Figueras A. 2004. Toxin and molecular analysis of Gymnodinium catenatum (Dinophyceae) strains from Galicia (NW Spain) and Andalucía (S Spain). J. Plankton Res. 26:341–349 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.