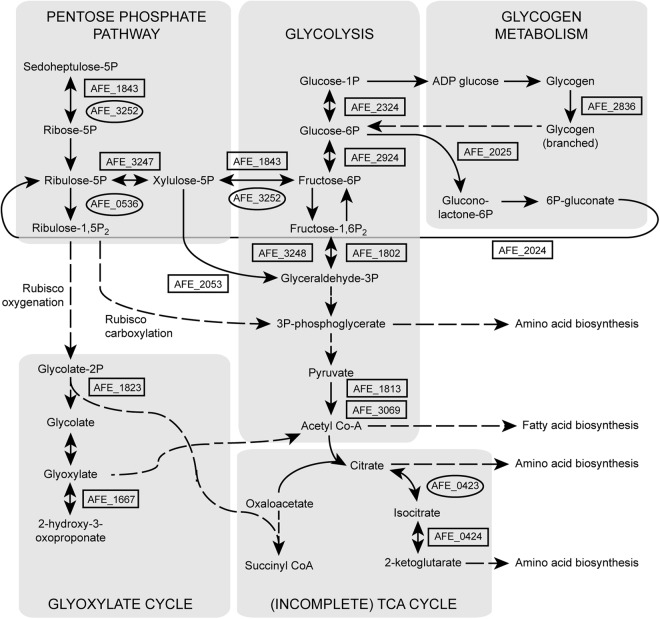
Fig 2.
Aerobic (gene numbers in boxes) and anaerobic (gene numbers in ovals) upregulated central carbon metabolism genes and proteins. Solid lines signify direct conversions, while dashed lines signify hidden intermediates (for clarity, not all connections have been included). The gene numbers refer to AFE_1843, tkt-1 transketolase (EC 2.2.1.1); AFE_3252, tkt-2 transketolase; AFE_3247, ribulose phosphate 3-epimerase; AFE_0536, phosphoribulokinase; AFE_2053, phosphoketolase; AFE_1823, phosphoglycolate phosphatase; AFE_1667, transketolase pyridine binding domain protein; AFE_2324, phosphoglucomutase; AFE_2924, glucose-6-phosphate isomerase (EC 5.3.1.9); AFE_1802, fructose-bisphosphate aldolase (class I); AFE_3248, fructose-bisphosphate aldolase (class II) (EC 4.1.2.13); AFE_3069, pyruvate dehydrogenase, E1 component, β subunit (EC 1.2.4.1); AFE_1813, dehydrogenase complex; AFE_2836, 1,4-α-glucan branching enzyme; AFE_2025, glucose-6-phosphate 1-dehydrogenase; AFE_2024, 6-phosphogluconate dehydrogenase family protein; AFE_0423, aconitate hydratase; AFE_0424, isocitrate dehydrogenase, NADP dependent. 5P, 5′-phosphate; 1,5-P2, 1,5-diphosphate; 2P, 2-phosphate; 1P, 1-phosphate; 6P, 6-phosphate; 1,6-P2, 1,6-diphosphate; 3P, 3′-phosphate; Acetyl Co-A, acetyl coenzyme A; 6P-gluconate, 6-phosphogluconate.