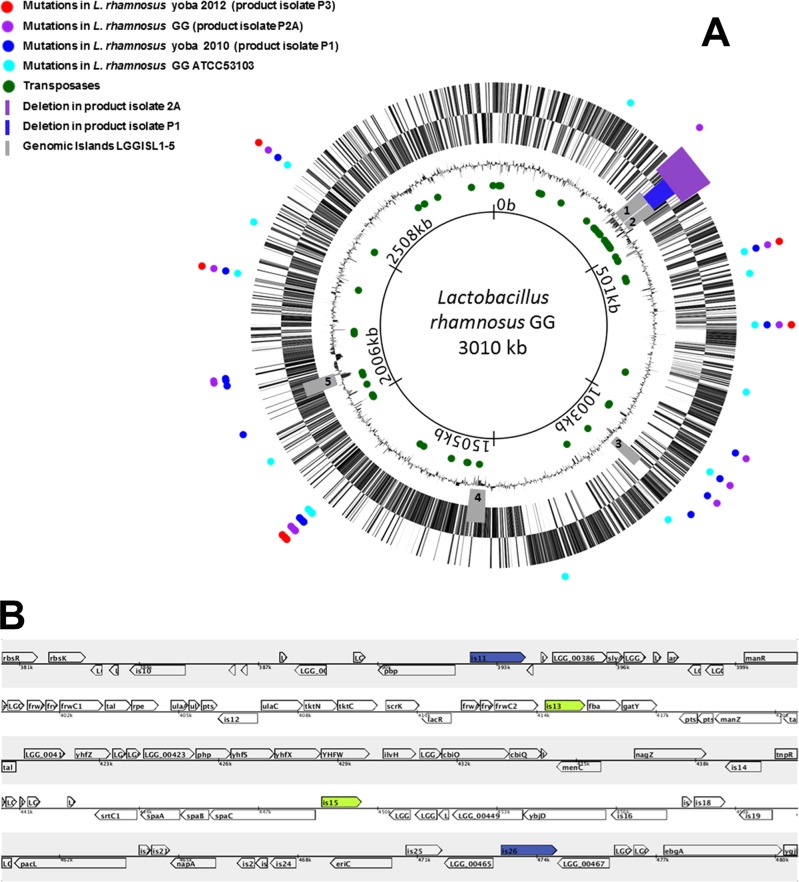
Fig 1.
(A) Circular genome map of L. rhamnosus GG (accession number NC_013198.1). Colored circles indicate the loci of the mutations in the ATCC 53103 reference strain and three probiotic product isolates including L. rhamnosus yoba 2010 and 2012. The purple and blue bars indicate the sites of the missing ORFs of isolates P2A and P1, respectively. In gray are the genomic islands LGGISL1 to LGGISL5. Circles indicate from outside to inside: genes for the “+” strand; genes for the “−” strand; the GC percentage; green circles, all IS elements encoding transposases in the genome; genome positions (total length, 3,010,111 bp). (B) Linear map showing the deletions including stretches of 34 genes flanked by two transposases of the IS30 family (is13 and is15 in green) in product isolate 1 and 84 genes flanked by two transposases of the IS5 family (is11 and is26 in blue) in product isolate 2A.