Fig 1.
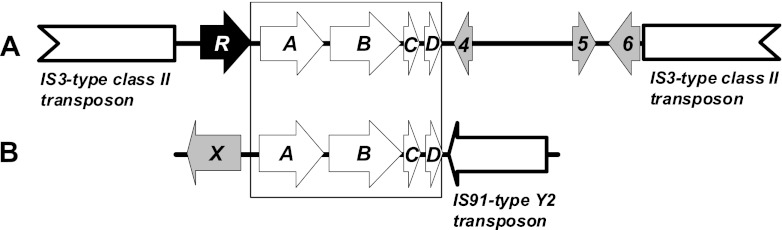
Comparison of the eth gene regions sequenced thus far in R. ruber IFP 2001 (20,138 bp, GenBank/EMBL/DDBJ database number AF333761) (A) and A. tertiaricarbonis L108 (6,909 bp, KC333876) (B), which encode fuel oxygenate ether monooxygenase Eth, consisting of ferredoxin reductase EthA, cytochrome P450 monooxygenase EthB, ferredoxin EthC, the unknown-function protein EthD, and other proteins. The box marks the DNA region showing 99.9% identical nucleotide sequences in the two strains. In contrast, strain IFP 2001 ethABCD genes are flanked by two identical copies of a 5.5-kb IS3-type class II transposon, the ethR gene, and other open reading frames (4 to 6), whereas the four eth genes in strain L108 are flanked by a 957-bp gene fragment (X) encoding a protein showing 99% sequence identity to the C-terminal part of a putative peptidyl-prolyl cis-trans isomerase from the MTBE-degrading strain Methylibium petroleiphilum PM1 (Mpe_A1955) and one copy of an 1.5-kb IS91-type Y2 transposon element.
