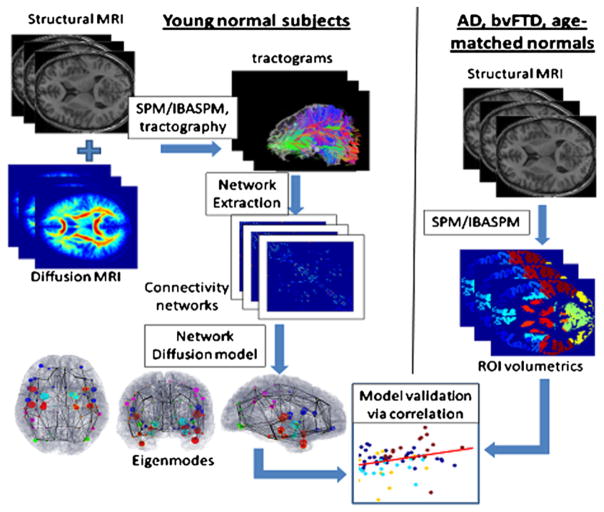
Figure 1. Process Diagram Depicting Our Methodology.
(Left) Structural healthy brain networks were obtained from diffusion MRI scans of 14 young healthy volunteers, followed by whole brain tractography. The nodes of this network correspond to cortical and subcortical gray matter regions obtained from a labeled T1-weighted brain atlas, and the edges of this network are proportional to the number and strength of the fiber tracts that connect the nodes. Proposed network diffusion model and its eigenmodes are derived from this healthy network. The first three eigenmodes, which we have hypothesized to be predictive of dementia atrophy patterns, are then tabulated and plotted.
(Right) We then compare the predicted patterns with measured atrophy of dementia patients (AD, bvFTD, and age-matched normal subjects), obtained via a completely separate processing pipeline, available in the SPM Matlab toolbox. T1-weighted images of each subject were coregistered with the same atlas as in the left panel, and gray-matter regions were parcellated using the prelabeled atlas information. Volume of each cortical and subcortical gray-matter region was measured. The atrophy of each region was obtained in terms of a t-statistic between the diseased and age-matched normal groups. Finally, the predicted and measured atrophy patterns were statistically compared using correlation analysis.