Abstract
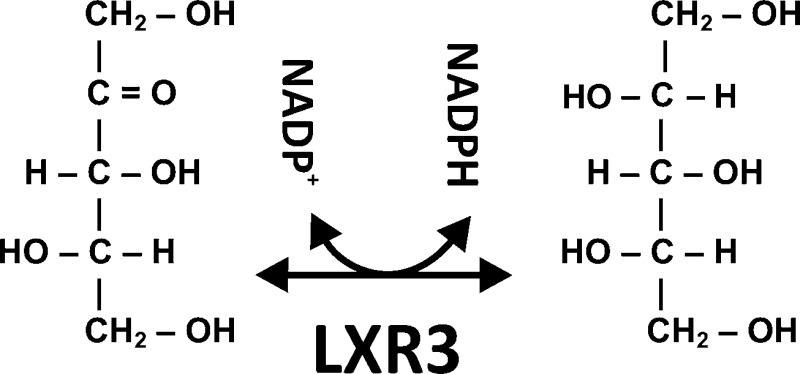
l-Xylulose reductases belong to the superfamily of short chain dehydrogenases and reductases (SDRs) and catalyze the NAD(P)H-dependent reduction of l-xylulose to xylitol in l-arabinose and glucuronic acid catabolism. Here we report the identification of a novel l-xylulose reductase LXR3 in the fungus Trichoderma reesei by a bioinformatic approach in combination with a functional analysis. LXR3, a 31 kDa protein, catalyzes the reduction of l-xylulose to xylitol via NADPH and is also able to convert d-xylulose, d-ribulose, l-sorbose, and d-fructose to their corresponding polyols. Transcription of lxr3 is specifically induced by l-arabinose and l-arabitol. Deletion of lxr3 affects growth on l-arabinose and l-arabitol and reduces total NADPH-dependent LXR activity in cell free extracts. A phylogenetic analysis of known l-xylulose reductases shows that LXR3 is phylogenetically different from the Aspergillus nigerl-xylulose reductase LxrA and, moreover, that all identified true l-xylulose reductases belong to different clades within the superfamily of SDRs. This indicates that the enzymes responsible for the reduction of l-xylulose in l-arabinose and glucuronic acid catabolic pathways have evolved independently and that even the fungal LXRs of the l-arabinose catabolic pathway have evolved in different clades of the superfamily of SDRs.
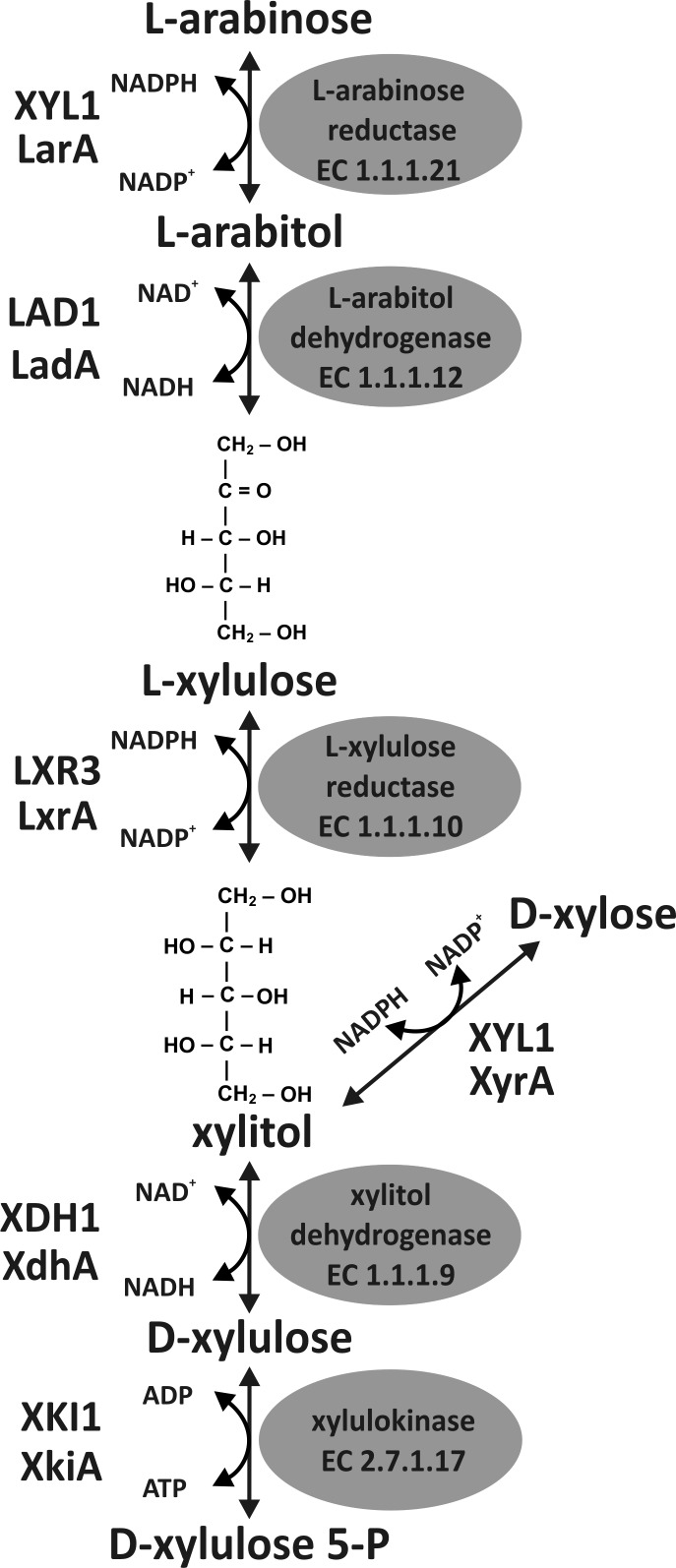
Plant cell walls consist of the polysaccharides cellulose, different hemicelluloses, and pectins and the complex polymer lignin. While cellulose is a linear β-1,4-linked d-glucose polymer, the structure and composition of hemicelluloses and pectins are more diverse. Following d-xylose, l-arabinose is the second most abundant pentose in hemicelluloses and pectins. It is present as a single residue or a short side chain in arabinoxylans or as larger branched side chains in the form of arabinan or arabinogalactan in pectins.1,2 Though the enzymatic steps for the catabolism of l-arabinose were described in the 1960s,3 most of the genes encoding the enzymes of this five-step pathway were only characterized recently.4 One reason for this might be that an l-arabinose pathway is not found in the fungal model organism Saccharomyces cerevisiae. Recently, considerable efforts were undertaken to fully elucidate this pathway for introduction of such a trait into S. cerevisiae to allow a complete conversion of plant biomass to, e.g., advanced biofuels or other biorefinery products.5,6 Degradation of l-arabinose in fungi usually consists of four oxidoreductive reactions and a final phosphorylation step, distinguishing this path from the different pathways for bacterial l-arabinose catabolism. The last two reactions of the fungal l-arabinose pathway are shared with the d-xylose catabolic pathway (Figure 1). The bacterial isomerase pathway consists of an l-arabinose isomerase, ribulokinase, and l-ribulose phosphate-4-epimerase, while the enzyme sequence of the oxidative pathway consists of l-arabinose dehydrogenase, l-arabinolactonase, l-arabonate dehydratase, l-2-keto-3-deoxy-arabonate dehydratase, and 2,5-dioxovalerate dehydrogenase, the end product being α-ketoglutarate. In a modification of this oxidative pathway, l-2-keto-3-deoxy-arabonate is split by an aldolase into pyruvate and glycoaldehyde.7,8 Most of the genes and proteins involved in the fungal l-arabinose pathway were characterized in the two ascomycetes Aspergillus niger and Trichoderma reesei.4 In T. reeseil-arabinose reduction is mediated by the NADPH specific d-xylose reductase XYL1, which is the major reductase activity for the reduction of both pentoses d-xylose and l-arabinose.9,10 In A. niger, this NADPH-dependent reduction is accomplished by an l-arabinose specific LarA and a d-xylose specific XyrA.11 The subsequent steps are mediated by l-arabitol 4-dehydrogenase,12,13l-xylulose reductase,14 xylitol dehydrogenase,15 and xylulose kinase.16
Figure 1.
Fungal l-arabinose degrading pathway represented by enzymes of A. niger and T. reesei. The first three specific steps of the fungal l-arabinose catabolism lead to xylitol, the first common intermediate of the l-arabinose and d-xylose pathway. Xylitol is then converted to d-xylulose 5-phosphate before entering the pentose phosphate pathway. l-Arabinose reduction is mainly mediated by the d-xylose reductase XYL1 in T. reesei, while A. niger has a specific l-arabinose reductase LarA.
Enzymes with l-xylulose reductase activity are found within the short chain dehydrogenase and reductase family17 and participate in the glucuronic acid/uronate cycle of mammals. In humans, LXR deficiency causes pentosuria, a clinically benign condition that results in large amounts of l-xylulose in the urine of such patients.18 The first fungal l-xylulose reductase, ALX1, was identified in the yeast Ambrosiozyma monospora and, interestingly, is NADH-dependent.19 Although an enzyme with l-xylulose reductase (LXR1) was described for T. reesei,20 its functional characterization showed that it is actually a d-mannitol 2-dehydrogenase.21,22 Only recently was a true l-xylulose reductase LxrA identified in A. niger. Its deletion resulted in an almost complete loss of the NADPH specific l-xylulose reductase activity but had an only small effect on the growth on l-arabinose as the carbon source, explained by the presence of a NADH-dependent l-xylulose reductase activity.14 However, deletion of the LxrA homologue LXR4 in T. reesei showed that this gene is not involved in the oxidoreductive catabolism of l-arabinose but of d-galactose.23
To clone putative LXRs involved in l-arabinose catabolism in T. reesei, we made use of the fact that all LXRs identified to date are found within the group of short chain dehydrogenases and reductases. Consequently, we screened the T. reesei genome database for SDRs encoding genes and reduced the number of LXR candidates by selecting for highly conserved fungal LXRs that are expressed in the presence of l-arabinose. Functional analysis identified a novel NADPH-dependent l-xylulose reductase that is involved in l-arabinose catabolism in T. reesei, which is different from the case for the previously described enzymes.
Materials and Methods
Strains and Growth Conditions
T. reesei QM9414 (ATCC 26921), Δlxr2, Δtku70,24 and Δlxr3 were cultivated on malt extract agar supplemented with uridine (10 mM) when necessary. Escherichia coli JM109 (Promega) was used for plasmid construction. For liquid cultivations, 106 spores per milliliter were incubated at 28 °C on a rotary shaker (250 rpm) in 250 mL of medium25 in 1 L Erlenmeyer flasks containing 1% (w/v) of the indicated carbon source. For replacement cultivations, strains were pregrown for 24 h with glycerol as the carbon source, washed with sterile media without the carbon source, and transferred to new medium with the indicated carbon source. Mycelia for biomass measurements were washed and dried to a constant weight at 80 °C. Dry biomass data are the average of three separate biological experiments with a deviation of <15%. Growth on solid substrates was recorded by inoculating agar plates with a piece of pregrown agar in the center and measuring the colony diameter daily.
Screening for T. reesei Putative l-Xylulose Reductase-Encoding Genes
One hundred genes encoding SDRs are found in the T. reesei genome database (http://genome.jgi-psf.org/Trire2/Trire2.home.html). Their corresponding protein sequences were used in a BLASTP search against the NCBI database to identify highly conserved proteins in mycelial fungi (e value of <10–80), followed by a BLASTP search to the genome database of the l-arabinose-utilizing yeast Candida guilliermondi (http://www.broadinstitute.org/annotation/genome/candida_guilliermondii; e value of <10–30). The number of candidate LXRs was then further reduced by selecting those genes for which respective ESTs were found in the NCBI T. reesei EST database. The GenBank entries of the other four genes of the l-arabinose pathway are CB905315.1 (xyl1), CF883445.1 (lad1), CF944055.1 (xdh1), and CF878255.1 (xki1). LXR3 was deposited as GenBank entry BK008567.
Construction of Fungal Strains
For deletion of lxr3, ∼1 kb of the lxr3 up- and downstream regions were amplified with specific primers (Table 1). The downstream region was ligated into pGEM-T Easy (Promega) followed by the SpeI/XhoI restricted upstream region and the SalI restricted orotidine-5′-monophosphate decarboxylase-encoding gene pyr4 as a selection marker,26 resulting in pBM1. A 4.9 kb NotI lxr3 deletion fragment was released from pBM1 and transformed into strain Δtku70 as described previously.26 For reintroduction of lxr3 into a Δlxr3 strain, the pyrithiamine resistance gene ptrA of Aspergillus oryzae was amplified from vector pME289227 with primers ptrA_fw_PstI and ptrA_rv_HindIII (Table 1) and ligated into pBluescript SK(+) (Stratagene). A 2.6 kb DNA fragment containing the whole lxr3 coding region, ∼1 kb of the upstream region, and 0.5 kb of the downstream region was amplified using the RElxr3-Acc65I/RElxr3-XhoI primer pair and introduced into the Acc65I and XhoI sites of this vector, resulting in pBM2. The Acc65I/HindIII fragment was used for transformation of Δlxr3 by electroporation.28 The reintroduction of lxr3 was verified by amplification of the 2.6 kb fragment by polymerase chain reaction (PCR) with oligonucleotides RElxr3-Acc65I and RElxr3-XhoI (data not shown). Deletion of lxr2 (tre54086) was described previously.24
Table 1. Oligonucleotides Used in This Study.
| oligonucleotide | sequence |
|---|---|
| ups-lxr3-Acc65I | GGTACCGTCTTCAACTCCTGATAGGG |
| ups-lxr3-XhoI | CTCGAGGGTCGGAGATCAAGAAAG |
| dws-lxr3-XhoI | CTCGAGCAACAGAAAGAGGTAGACC |
| dws-lxr3-XbaI | TCTAGACAACTTTAGCACCTGGAGC |
| RElxr3-Acc65I | GGTACCAACTCCTCGACCGAAATAG |
| RElxr3-XhoI | CTCGAGTCATGCTCATTGTGTGCTCC |
| ptrA_fw_PstI | TCTGCAGAAAGCTAGGAGATCGTCC |
| ptrA_rv_HindIII | TAAGCTTCTCTTGCATCTTTGTTTG |
| rc_lxr3_HisN_fw_EcoRI | ATATGAATTCACAATGCATCACCATC |
| ACCATCACGGGAAGAACGGCGCCTTTCCG | |
| rc_lxr3_rv_EcoRV | TAATGATATCTCATGGCAGGCTGTAGCCGCC |
| qPCR_tef1_fw | CCACATTGCCTGCAAGTTCGC |
| qPCR_tef1_rv | GTCGGTGAAAGCCTCAACGCAC |
| qPCR_xyl1_fw | AGAACCTGGACAACACCTC |
| qPCR_xyl1_rv | GGCGGAGAAGTAGTTTGTAG |
| qPCR_lad1_fw | GAGCGGTGTCATCGATCTATC |
| qPCR_lad1_rv | TCTTGGGATCTGCTGACGTCTC |
| qPCR_lxr3_fw | AACAGCTCCAAGGCCGCCGTGATTC |
| qPCR_lxr3_rv | AGACACGGTGTTGACGCGGGCAAAG |
| qPCR_xdh1_fw | GCATCTCGGCTGAGGACAAC |
| qPCR_xdh1_rv | CGTGAATGCTCGTCTGGATC |
| qPCR_lxr2_fw | GCCGATATTGGAACAGACG |
| qPCR_lxr2_rv | GAAGACTGCGCCAATGTAC |
| qPCR_tre122079_fw | TCCAAGGCTGGTGTCATGC |
| qPCR_tre122079_rv | ATCCAGGCGAGAGTGTGTTG |
Nucleic Acid Isolation and Transcriptional Analysis
Fungal mycelia were harvested by filtration, washed with cold tap water, frozen, and ground in liquid nitrogen. Following RNA isolation,29 5 μg of total RNA was treated with DNase [DNase I, RNase free (Fermentas)] and reverse transcribed [RevertAid First Strand cDNA Kit (Fermentas)] using a 1:1 mixture of oligo-dT and random hexamer primers. To test for potential LXR-encoding genes, reverse transcription PCR (RT PCR) was performed with RNA isolated from T. reesei QM9414. Strain QM9414 was pregrown on medium containing glycerol as the carbon source followed by a transfer to new medium with l-arabinose, d-galactose, or d-glucose [1% (w/v)] as the carbon source. Data are found in Table S1 of the Supporting Information. Quantitative real-time PCRs (qPCRs) were performed by the iCycler iQ real-time detection system (Bio-Rad). Each reaction mixture contained 1 μL of the 1:10 diluted cDNA (approximately 2.5 ng), 12.5 μL of the iQ SYBR Green Supermix (Bio-Rad), primers (Table 1, final concentration of 100 nM), and nuclease free water in a final volume of 25 μL. Primer efficiency was calculated using a dilution series from 1:1 to 1:1000 with the PCR baseline-subtracted mode. The threshold cycles (CT) were adjusted for an optimal efficiency of 2. The amplification protocol consisted of an initial denaturation step for 3 min at 95 °C followed by 40 cycles of denaturation (95 °C for 15 s), annealing, and elongation (61 °C for 20 s). qPCRs were conducted in triplicate. Data calculation was performed with iQ5 Optical System software version 2.0 (Bio-Rad) and REST.30 Individual samples were normalized to the expression of tef1 (translation elongation factor 1α) as described previously.31
Phylogenetic Analysis
Phylogenetic analysis was performed using CLUSTALX version 1.832 for protein sequence alignment, GENEDOC version 2.633 for visual adjustment, and MEGA version 534 for construction of phylogenetic trees. Neighbor joining was used as the algorithm for distance calculation and evaluated by 1000 bootstrap rearrangements. To retrieve closely related SDR sequences from other species, T. reesei candidate SDRs were used in a BLASTP search against the NCBI database.
Recombinant Production and Purification of LXR3
Expression, protein extraction, and purification of LXR3 in S. cerevisiae strain CEN.PK2-1D (European S. cerevisiae Archive for Functional Analysis) were performed as described for A. niger LxrA.14 An lxr3 cDNA was amplified with primers rc_lxr3_HisN_fw_EcoRI and rc_lxr3_rv_EcoRV (Table 1) and cloned in pYX212 [URA3 selection (Ingenius R&D Systems, Madison, WI)], allowing expression of lxr3 under the TPI1 (triosephosphate isomerase) promoter to produce the recombinant LXR3 with an N-terminal His tag. The lxr3 cDNA was verified by sequencing.
Preparation of T. reesei Cell Free Extracts
T. reesei mycelia grown in a liquid culture were washed and ground in liquid nitrogen. Per gram of mycelia (wet biomass) 3 mL of extraction buffer was added (PBS) [8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4 (pH 7.4), and 5 mM β-mercaptoethanol] and the mixture homogenized (12 × 20 s, duty cycle of 25%, output of 2) with a Branson model 250 sonifier at 4 °C. After centrifugation (10000 rpm for 10 min at 4 °C), 20% glycerol (final concentration) was added and the cell free extracts were stored at −80 °C.
For the LXR activity measurements of T. reesei grown in the rich medium, 100 mL of YPG medium containing 10 g/L yeast extract, 2 g/L Bacto peptone, and 3% Difco gelatin (Becton Dickinson and Co.) was inoculated with 1 mL of the spore suspension. Overnight (16 h) growth at 28 °C resulted in a dense homogeneous mycelium suspension, which was collected by filtration and split into two comparable portions. The mycelia were resuspended in 50 mL of YP medium supplemented with either 1% d-glucose or 1% l-arabinose and incubated for 6 h at 28 °C. For LXR activity measurements on minimal medium, 100 mL of medium containing 1% (w/v) glycerol was incubated for 24 h, and the mycelia were collected by filtration, split into two comparable portions, resuspended in 50 mL of MM medium supplemented with either 1% d-glucose or 1% l-arabinose, and incubated for 15 h at 28 °C. Following induction, mycelia were isolated by filtration and washed with water, and an appropriate amount of mycelia was transferred to a 2 mL tube with 0.6 mL of acid-washed glass beads (Sigma), 1 mL of lysis buffer [500 mM NaCl and 50 mM NaH2PO4 (pH 8.0)], and protease inhibitors (Complete, Roche). The cells were disrupted in a 30 s breaking session in a Precellys 24 instrument (Bertin Technologies). The cell extracts were clarified by centrifugation, and the supernatants were used in the enzyme assays. The protein concentration was measured using the Protein Assay kit (Bio-Rad).
Enzyme and Polyol Assays
The enzyme activity of cell free extracts was measured with a NanoPhotometer Pearl (Implen) or Helios Beta UV–vis spectrophotometer (Thermo Scientific) by recording the rate of change in absorbance at 340 nm for NAD(P)+ reduction and NAD(P)H oxidation. Polyol oxidation was performed in 100 mM Tris-HCl (pH 9.0) and 2 mM NAD(P)+ in the presence of 100 μg of cell free extracts and started with addition of 100 mM substrate. For sugar reduction, 100 mM HEPES-NaOH (pH 7.0) and 0.2 mM NAD(P)H were used.
Enzyme activity measurements of recombinantly produced proteins were performed by varying the substrate concentration over the range of 5–285 mM in 50 mM Tris-HCl buffer (pH 7.0) with 0.5 mM NADPH for sugar reduction and 100 mM Tris-HCl (pH 8.0) with 1 mM NADP+ for polyol oxidation. For analysis of the kinetic constants with NADPH, the activity was measured with varying NADPH concentrations over the range of 8–500 μM in 50 mM Tris-HCl buffer (pH 7.0) with 125 mM l-xylulose. Reactions were initiated by addition of the enzyme. The different substrate concentrations are indicated in the results. Enzyme assays were performed in microtiter plates (NUNC) with a Varioscan spectrophotometer (Thermo Electron Corp.). Activities are expressed in nanokatals and are given as specific activities (nanokatal per milligram of protein). High-performance liquid chromatography measurements were performed as described previously.10,21 Enzyme measurements for l-arabinose reductase, l-arabitol dehydrogenase, and xylitol dehydrogenase activity in cell free extracts were described previously.35,36
Results
Identification of Putative T. reeseil-Xylulose Reductases
All l-xylulose reductases characterized to date belong to the superfamily of short chain dehydrogenases and reductases (SDR). We therefore screened the T. reesei genome database for genes encoding putative LXRs and identified ∼117 different SDRs. To reduce the number of putative candidate LXRs, we reduced their number by presuming the following: an l-xylulose reductase is a highly conserved enzyme, and therefore, orthologues should be present in the genomes of most mycelial fungi and present in the l-arabinose-utilizing yeast C. guilliermondi. Because the genes encoding the other four steps of the l-arabinose pathway in T. reesei are represented by ESTs in the NCBI database, we also tested if our potential LXRs are present in this EST database. To further reduce the number for functional analysis, we tested their expression by RT PCR under l-arabinose inducing conditions and compared it to their expression on d-glucose. For seven genes, we found transcription under all conditions, but only three were specifically induced by l-arabinose. Because we originally also assumed that an LXR would be induced by d-galactose, we chose the two genes that showed on both sugars induction and termed them lxr2 (tre54086) and lxr3 (tre60033). An overview of the results of the in silico and expression analysis of 20 candidates is given in Table S1 of the Supporting Information.
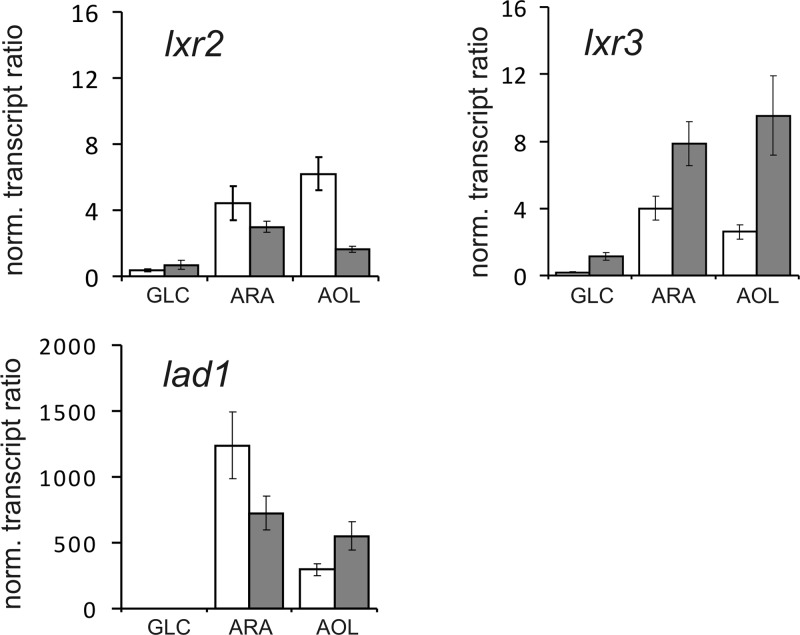
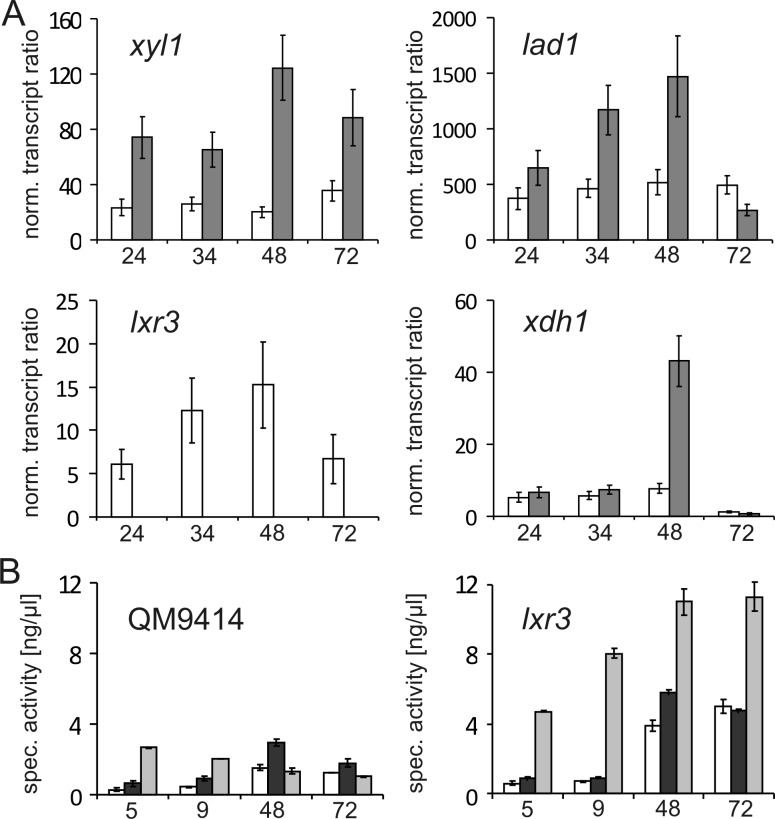
Their transcriptional response to the presence of different inducers was then quantified by qPCR using lad1 as a positive control for an l-arabinose inducible gene (Figure 2). lxr3 showed increased transcript levels when induced by l-arabinose and l-arabitol and an increase in transcript level from 2 to 8 h after replacement. lxr2 also exhibited upregulation with highest transcript levels found at the earlier time point on l-arabinose or l-arabitol. In comparison to both lxr2 and lxr3, lad1 showed a higher inducibility on both l-arabinose and l-arabitol, which is due to its lower basal transcription level on glycerol.
Figure 2.
Transcriptional analysis of T. reesei lxr2 and lxr3. T. reesei QM9414 was cultivated for 24 h on glycerol and replaced with new medium containing 1% (w/v) of the indicated carbon source (GLC, d-glucose; ARA, l-arabinose; AOL, l-arabitol) for 2 (gray bars) and 8 h (white bars). Expression of lxr2, lxr3, and lad1 is related to their expression on glycerol after 24 h and normalized to the expression of tef1.
Effect of Deletion of lxr2 and lxr3 on Growth
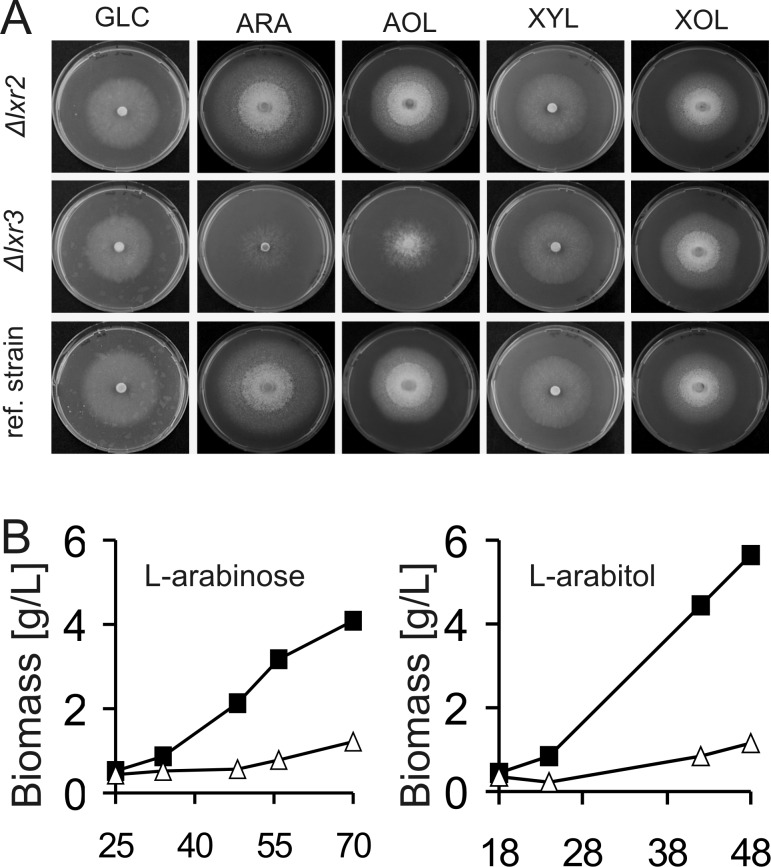
To test the potential role of lxr3 in fungal l-arabinose catabolism, we produced a knockout cassette for lxr3 in which the lxr3 coding region was replaced by the T. reesei pyr4 marker gene. Following transformation and analysis of the purified transformants by diagnostic PCR, several lxr3 deletion strains were identified. The growth behavior of the Δlxr224 and Δlxr3 strains was tested on different carbon sources. In this test, Δlxr2 strains showed no specific growth phenotype compared to its parental strain (Figure 3A). This was in contrast to Δlxr3 strains: here levels of growth on solid medium and biomass accumulation during liquid cultivation were strongly decreased for both l-arabinose and l-arabitol (Figure 3A,B). No effect, however, was found for growth with, e.g., d-glucose or d-xylose as the carbon source. A reintroduction of lxr3 into the Δlxr3 strain restored growth on l-arabinose and l-arabitol (Figure 1 of the Supporting Information).
Figure 3.
Effect of deletion of lxr2 and lxr3 on growth on different carbon sources. (A) Radial growth on agar plates after 3 days and (B) biomass accumulation during liquid cultivation on different carbon sources (1%, w/v) as indicated for lxr3 (△) compared to the parental strain (■): GLC, d-glucose; ARA, l-arabinose; AOL, l-arabitol; XYL, d-xylose; XOL, xylitol.
Deletion of lxr3 Affects the Total l-Xylulose Activity and the Regulation of l-Arabinose Metabolism
The prominent effect of the lxr3 deletion on the utilization of the carbon sources l-arabinose and l-arabitol was further investigated by determining the total l-xylulose reductase activity produced in cell free extracts in the Δlxr3 strain. l-Arabinose-induced cell free extracts were prepared from mycelia after replacement to minimal medium as well as rich medium with l-arabinose as the inducing carbon source. Deletion of lxr3 led to a significant reduction in NADPH specific LXR activity after replacement to both media containing l-arabinose (Figure 4), while NADH specific LXR activity remained constant (e.g., 0.6 nkat/mg on l-arabinose-containing minimal medium). Again, the deletion of lxr2 had no negative influence on LXR activity, indicating that LXR3 is responsible for the major NADPH specific l-xylulose reductase in T. reesei during growth on l-arabinose as the carbon source. To examine if a deletion of lxr3 has an influence on other genes involved in the l-arabinose catabolism, we performed further transcriptional studies. This analysis shows that the transcript levels of xyl1 and lad1 are considerably upregulated in the Δlxr3 strain during the whole cultivation period compared to that of the reference strain, while upregulation of xdh1 is found only at a later time point around 48 h (Figure 5A). This change in the transcription profile was also reflected by the elevated total enzyme activities for l-arabinose reductase, l-arabitol dehydrogenase, and xylitol dehydrogenase in the cell free extracts (Figure 5B).
Figure 4.
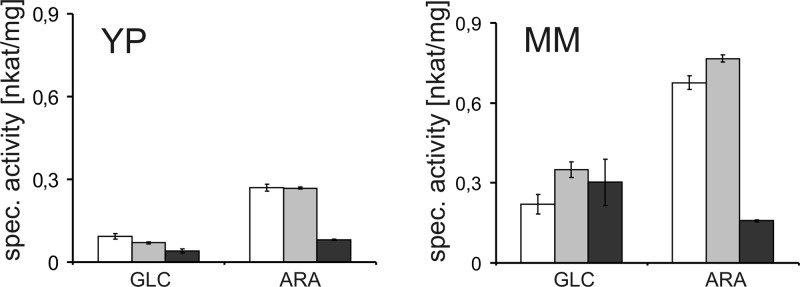
Effect of the deletion of lxr2 and lxr3 on total l-xylulose reductase activities. Mycelia were pregrown before the medium was replaced with rich (YP) or minimal medium (MM) containing either 1% (w/v) d-glucose or l-arabinose for 6 or 15 h. NADPH-dependent LXR activity was tested in crude protein extracts for QM9414 (white bars), Δlxr2 (gray bars), and Δlxr3 (dark gray bars).
Figure 5.
Consequences of the deletion of lxr3 on the expression of other genes of the l-arabinose pathway. (A) Transcript levels of xyl1, lad1, xdh1, and lxr3 relative to the expression during growth on glycerol at 24 h and normalized to tef1. (B) Total l-arabinose reductase (white bars), l-arabitol dehydrogenase (dark gray bars), and xylitol dehydrogenase (light gray bars) activities were measured in QM9414 and Δlxr3. Strains were either precultivated on glycerol and replaced with l-arabinose (5 and 9 h) or cultivated directly on l-arabinose (48 and 72 h).
Characterization of T. reesei LXR3
Functional analysis supports the role of LXR3 in l-arabinose catabolism. To characterize the enzyme with respect to its l-xylulose reductase activity, we expressed LXR3 recombinantly in S. cerevisiae and investigated substrate specificities and enzyme kinetics. The purified enzyme reduced l-xylulose with a Km of 16 mM, a Vmax of 367 nkat/mg, and a kcat of 11.4 s–1. For NADPH, we obtained a Km of 0.13 mM, a Vmax of 250 nkat/mg, and a kcat of 7.75 s–1.
LXR3 also exhibited activity with d-ribulose (Km = 105 mM; Vmax = 266 nkat/mg; kcat = 8.24 s–1) and with polyols d-sorbitol (Km = 250 mM; Vmax = 58 nkat/mg; kcat = 1.8 s–1) and xylitol (Km = 100 mM; Vmax = 33 nkat/mg; kcat = 1 s–1) and weak activity with d-xylulose, l-sorbose, and d-fructose (Vmax < 30 nkat/mg). No activity was recorded with l-xylo-3-hexulose, the substrate of LXR4 in the oxidoreductive d-galactose pathway,23d-sorbose, d-ribitol, d-arabitol, or l-arabitol. The enzyme was also strictly NADP(H) specific, and no activity was observed with NADH as the cosubstrate.
Phylogenetic Analysis of l-Xylulose Reductases
The fact that T. reesei LXR3 is quite dissimilar from A. niger LxrA, while both are in vivo functional l-xylulose reductases, prompted us to investigate their phylogenetic relationship. We also included the other three T. reesei LXR proteins, LXR1 (d-mannitol 2-dehydrogenase, which also exhibits l-xylulose reductase activity), LXR2, and LXR4 (l-xylo-3-hexulose reductase), and used them as a query in a BLASTP search against the NCBI database. The resulting best hits were pruned from duplicates, and 182 protein sequences were subjected to a neighbor joining analysis and rooted to the corresponding ALX1 from Am. monospora and two other proteins from different yeasts. The result (Figure 6 and Figure 2 of the Supporting Information) shows that the fungal LXR proteins form three major clades: one basal clade leading to a large clade that contained the A. niger functional l-xylulose reductase LxrA and the l-xylo-3-hexulose reductase LXR4, another that contained LXR1, and a third that was split into two subclades containing LXR2 and LXR3. Branches within the LXR3 clade showed poor bootstrap support and displayed paralogs in many species, particularly Pyrenomycetes (also Trichoderma virens and Trichoderma atroviride but not T. reesei). This suggests that l-xylulose reductases and related enzymes have proliferated in Pyrenomycetes and thereby apparently adapted their substrate specificity. From this analysis, it is obvious that the trait for l-xylulose reductase has evolved independently within the family of short chain dehydrogenases for enzymes of the l-arabinose pathway and the glucuronic acid pathway and that even the fungal LXRs involved in l-xylulose reduction in the l-arabinose catabolic pathway have evolved in different clades of SDRs.
Figure 6.
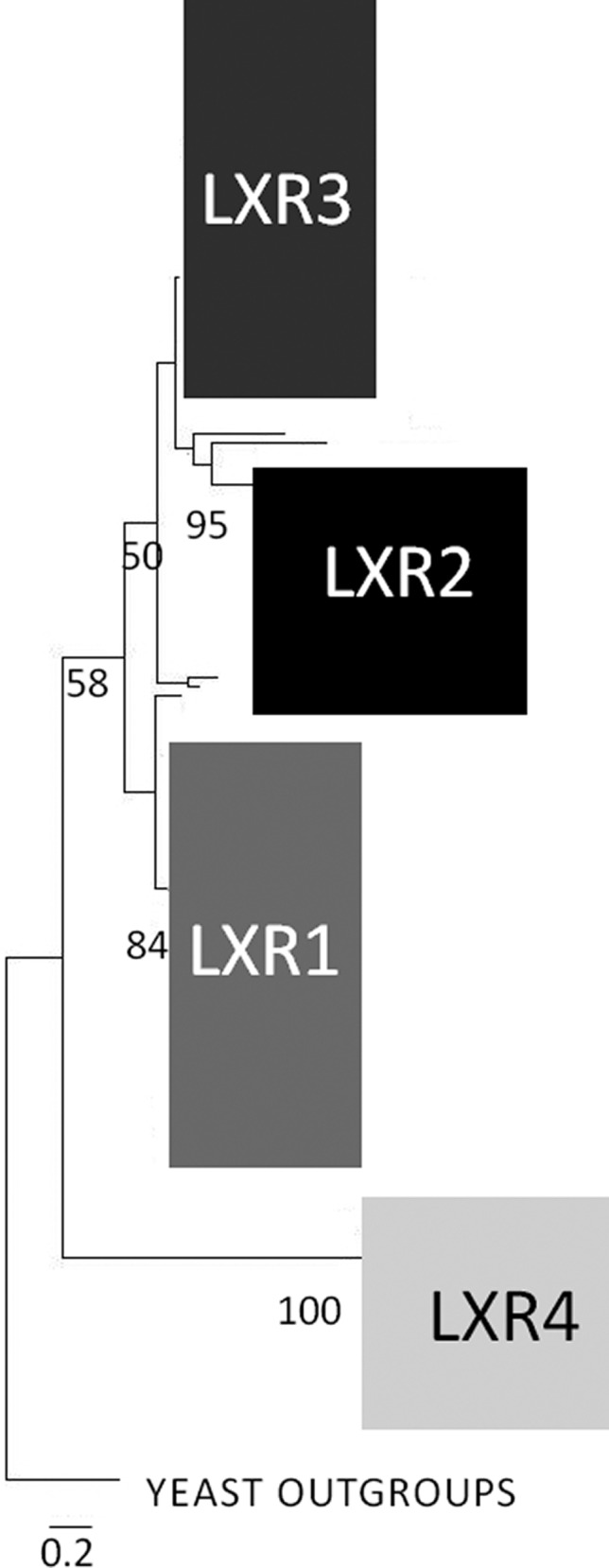
Scheme of the phylogenetic relationship of T. reesei LXR3 to other in vivo functional l-xylulose reductases, including the l-xylulose reductase LxrA of A. niger and ALX1 of Am. monospora found in yeast outgroups. Also included are LXR1 (d-mannitol 2-dehydrogenase, which also exhibits l-xylulose reductase activity), LXR2, and the l-xylo-3-hexulose reductase LXR4. The numbers below nodes indicate the bootstrap value. The bar marker indicates the genetic distance, which is proportional to the number of amino acid substitutions. The detailed phylogenetic tree is found in Figure 2 of the Supporting Information.
Discussion
Most of the genes and their corresponding enzymes involved in the l-arabinose and d-xylose pathway have been characterized from the two ascomycetes A. niger and T. reesei. Although the overall sequence of reactions consisting of four oxidoreductive steps and a final phosphorylation is conserved (Figure 1), a comparative functional analysis reveals a number of species specific adaptations such as the presence of a single enzyme (XYL1) for both l-arabinose and d-xylose reduction in T. reesei, but two rather specific reductases for l-arabinose (LarA) and d-xylose (XyrA) in A. niger.11,36 Here we identify a further difference and show that T. reesei uses a novel l-xylulose reductase LXR3 in l-arabinose catabolism.
In the past, different enzymes responsible for l-xylulose reduction were identified in the family of SDRs. The first enzymes responsible for l-xylulose reduction were found in mammals, where their absence blocked the pathway for d-glucuronic acid leading to the accumulation of l-xylulose in blood and urine. The molecular background of this condition, pentosuria, was recently elucidated.18 To date, two fungal LXRs have been functionally verified to be involved in l-arabinose catabolism, including the NADH-dependent ALX1 of the yeast Am. monospora(19) and the recently identified NADPH-dependent LxrA of A. niger.14 Although it was assumed for many years that the LXR step is NADPH-dependent,3,38 recent investigations in A. niger showed that the NADPH-dependent l-xylulose reductase activity is not needed for rapid growth on l-arabinose. The situation is obviously different in T. reesei where the strongly reduced NADPH-dependent l-xylulose activity as a consequence of lxr3 deletion leads to a severe reduction in the level of growth on l-arabinose and l-arabitol as carbon sources. The question of whether the NADH-dependent activity in T. reesei is too low to replace the NADPH-dependent activity as suggested for A. niger remains.14,38 Future research will clarify if a NADH-dependent LXR is responsible for the major conversion of l-xylulose to xylitol in A. niger. A NADH-dependent l-xylulose reductase step would also have consequences for the redox balance. Although the overall process of l-arabinose assimilation is redox neutral, it leads to an unequal use of cofactors with two reductive NADPH-dependent and two oxidative NAD+-dependent steps. Interestingly, the Vmax of purified LXR3 is much lower than that of purified A. niger LxrA (Vmax = 10833.3 nkat/mg)14 but higher than that of purified LXR1 (75 nkat/mg).20
The l-xylulose reductases and related proteins appear to have undergone an intriguing evolution: the presence of members of Eurotiomycetes, Dothidiomycetes, and Pyrenomycetes in both large phylogenetic clades suggests that there have been early duplication events that were followed by gene losses in the former two classes with the exception of the Pyrenomycetes where the gene losses were less intense. In addition, the substrate specificity of these proteins appears to have undergone readjustment as, e.g., LXR3 is not able to convert l-xylo-3-hexulose whereas LXR4 is able to do so.23 As a consequence of this evolution, T. reeseil-xylulose reductase LXR3 is more closely related to T. reeseid-mannitol dehydrogenase LXR1 than to A. nigerl-xylulose reductase LxrA. Originally, we assumed that LXR3 might also be responsible for the conversion of l-xylo-3-hexulose, the product of LAD1,13 to the corresponding polyol d-sorbitol in the oxidoreductive d-galactose pathway. This would be analogous to the findings that in T. reesei other l-arabinose pathway enzymes such as XYL1 and LAD1 function in this oxidoreductive d-galactose catabolism. However, our results show that LXR3 is not able to convert l-xylo-3-hexulose. We have recently identified yet another SDR LXR4 that is involved in this step in oxidoreductive d-galactose catabolism.23
A major consequence of the lxr3 deletion is the disturbance of l-arabinose catabolism. Its deletion results in a specific upregulation of genes of the l-arabinose pathway acting upstream of lxr3, i.e., xyl1 and lad1, while xdh1 that is responsible for the step downstream of lxr3 is only upregulated to a later time point. This would imply that the inducer for l-arabinose catabolic genes xyl1 and lad1 is produced upstream of lxr3, while the inducer for the upregulation of xdh1 accumulates at a later time point..
Acknowledgments
We thank Martina Marchetti-Deschmann for providing an Implen Nanophotometer.
Glossary
Abbreviations
- SDRs
short chain dehydrogenases and reductases
- LXR
l-xylulose reductase
- ESTs
expressed sequence tags.
Supporting Information Available
Overview of the in silico and expression analysis of the 20 candidates (Table S1), the complementation of lxr3 in a Δlxr3 strain (Figure 1), and a detailed phylogenetic tree of different LXR proteins (Figure 2). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ B.M.: Department of Biotechnology, Delft University of Technology and Kluyver Centre for Genomics of Industrial Fermentation, Julianalaan 67, 2628 BC Delft, The Netherlands.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Author Contributions
D.M. and S.H. contributed equally to this work.
This study was supported by the Austrian Science Fund FWF to B.S. (P19421) and Grant 131869 of the Academy of Finland. S.H. is a member of the Ph.D. school program Applied Bioscience Technologies AB-Tec financed by Vienna University of Technology. S.H. is also supported by the Austrian Science Fund FWF (P24219).
The authors declare no competing financial interest.
Supplementary Material
References
- Gilbert H. J. (2010) The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153, 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil M. A., and York W. S. (2003) The composition and structure of plant primary walls. In The Plant Cell Wall (Rose J. K. C., Ed.) pp 1–54, Blackwell, Oxford, U.K. [Google Scholar]
- Chiang C.; Knight S. G. (1961) l-Arabinose metabolism by cell-free extracts of Penicillium chrysogenum. Biochim. Biophys. Acta 46, 271–278. [DOI] [PubMed] [Google Scholar]
- Seiboth B.; Metz B. (2011) Fungal arabinan and l-arabinose metabolism. Appl. Microbiol. Biotechnol. 89, 1665–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P.; Verho R.; Putkonen M.; Londesborough J.; Penttilä M. (2003) Production of ethanol from l-arabinose by Saccharomyces cerevisiae containing a fungal l-arabinose pathway. FEMS Yeast Res. 3, 185–189. [DOI] [PubMed] [Google Scholar]
- Hahn-Hägerdal B.; Karhumaa K.; Jeppsson M.; Gorwa-Grauslund M. F. (2007) Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108, 147–177. [DOI] [PubMed] [Google Scholar]
- Watanabe S.; Shimada N.; Tajima K.; Kodaki T.; Makino K. (2006) Identification and characterization of l-arabonate dehydratase, l-2-keto-3-deoxyarabonate dehydratase, and l-arabinolactonase involved in an alternative pathway of l-arabinose metabolism. Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 281, 33521–33536. [DOI] [PubMed] [Google Scholar]
- Schleif R. (2000) Regulation of the l-arabinose operon of Escherichia coli. Trends Genet. 16, 559–565. [DOI] [PubMed] [Google Scholar]
- Seiboth B.; Gamauf C.; Pail M.; Hartl L.; Kubicek C. P. (2007) The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for β-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66, 890–900. [DOI] [PubMed] [Google Scholar]
- Akel E.; Metz B.; Seiboth B.; Kubicek C. P. (2009) Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryotic Cell 8, 1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzita D.; Penttilä M.; Richard P. (2010) Identification of an l-arabinose reductase gene in Aspergillus niger and its role in l-arabinose catabolism. J. Biol. Chem. 285, 23622–23628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P.; Londesborough J.; Putkonen M.; Kalkkinen N.; Penttilä M. (2001) Cloning and expression of a fungal l-arabinitol 4-dehydrogenase gene. J. Biol. Chem. 276, 40631–40637. [DOI] [PubMed] [Google Scholar]
- Pail M.; Peterbauer T.; Seiboth B.; Hametner C.; Druzhinina I.; Kubicek C. P. (2004) The metabolic role and evolution of l-arabinitol 4-dehydrogenase of Hypocrea jecorina. Eur. J. Biochem. 271, 1864–1872. [DOI] [PubMed] [Google Scholar]
- Mojzita D.; Vuoristo K.; Koivistoinen O. M.; Penttilä M.; Richard P. (2010) The ‘true’ l-xylulose reductase of filamentous fungi identified in Aspergillus niger. FEBS Lett. 584, 3540–3544. [DOI] [PubMed] [Google Scholar]
- Seiboth B.; Hartl L.; Pail M.; Kubicek C. P. (2003) d-Xylose metabolism in Hypocrea jecorina: Loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded l-arabinitol-4-dehydrogenase. Eukaryotic Cell 2, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanKuyk P. A.; de Groot M. J.; Ruijter G. J.; de Vries R. P.; Visser J. (2001) The Aspergillus nigerd-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on d-xylose and l-arabinose. Eur. J. Biochem. 268, 5414–5423. [DOI] [PubMed] [Google Scholar]
- Kallberg Y.; Oppermann U.; Jornvall H.; Persson B. (2002) Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269, 4409–4417. [DOI] [PubMed] [Google Scholar]
- Pierce S. B.; Spurrell C. H.; Mandell J. B.; Lee M. K.; Zeligson S.; Bereman M. S.; Stray S. M.; Fokstuen S.; MacCoss M. J.; Levy-Lahad E.; King M. C.; Motulsky A. G. (2011) Garrod’s fourth inborn error of metabolism solved by the identification of mutations causing pentosuria. Proc. Natl. Acad. Sci. U.S.A. 108, 18313–18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verho R.; Putkonen M.; Londesborough J.; Penttilä M.; Richard P. (2004) A novel NADH-linked l-xylulose reductase in the l-arabinose catabolic pathway of yeast. J. Biol. Chem. 279, 14746–14751. [DOI] [PubMed] [Google Scholar]
- Richard P.; Putkonen M.; Vaananen R.; Londesborough J.; Penttilä M. (2002) The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41, 6432–6437. [DOI] [PubMed] [Google Scholar]
- Metz B.; de Vries R. P.; Polak S.; Seidl V.; Seiboth B. (2009) The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a d-mannitol dehydrogenase and is not involved in l-arabinose catabolism. FEBS Lett. 583, 1309–1313. [DOI] [PubMed] [Google Scholar]
- Metz B.; Seidl-Seiboth V.; Haarmann T.; Kopchinskiy A.; Lorenz P.; Seiboth B.; Kubicek C. P. (2011) Expression of biomass-degrading enzymes is a major event during conidium development in Trichoderma reesei. Eukaryotic Cell 10, 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzita D.; Herold S.; Metz B.; Seiboth B.; Richard P. (2012) l-Xylo-3-hexulose reductase is the missing link in the oxidoreductive pathway for d-galactose catabolism in filamentous fungi. J. Biol. Chem. 287, 26010–26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guangtao Z.; Hartl L.; Schuster A.; Polak S.; Schmoll M.; Wang T.; Seidl V.; Seiboth B. (2009) Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J. Biotechnol. 139, 146–151. [DOI] [PubMed] [Google Scholar]
- Mandels M. M.; Andreotti R. E. (1978) The cellulose to cellulase fermentation. Proc. Biochem. 13, 6–13. [Google Scholar]
- Gruber F.; Visser J.; Kubicek C. P.; de Graaff L. H. (1990) The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18, 71–76. [DOI] [PubMed] [Google Scholar]
- Krappmann S.; Bayram O.; Braus G. H. (2005) Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryotic Cell 4, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A.; Bruno K. S.; Collett J. R.; Baker S. E.; Seiboth B.; Kubicek C. P.; Schmoll M. (2012) A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P.; Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W.; Horgan G. W.; Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V.; Druzhinina I. S.; Kubicek C. P. (2006) A screening system for carbon sources enhancing β-N-acetylglucosaminidase formation in Hypocrea atroviridis (Trichoderma atroviride). Microbiology 152, 2003–2012. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., and Higgins D. G. (2002) Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics, Chapter 2, Unit 2, 3, Wiley, New York. [DOI] [PubMed] [Google Scholar]
- Nicholas H. B. Jr.; McClain W. H. (1987) An algorithm for discriminating sequences and its application to yeast transfer RNA. Comput. Appl. Biosci. 3, 177–181. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Nei M.; Dudley J.; Tamura K. (2008) MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinf. 9, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B.; Hartl L.; Pail M.; Kubicek C. P. (2003) d-Xylose metabolism in Hypocrea jecorina: Loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded l-arabinitol-4-dehydrogenase. Eukaryotic Cell 2, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B.; Gamauf C.; Pail M.; Hartl L.; Kubicek C. P. (2007) The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for β-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66, 890–900. [DOI] [PubMed] [Google Scholar]
- Witteveen C. F. B.; Busink R.; van de Vondervoort P. J.; Dijkema C.; Swart K.; Visser J. (1989) l-Arabinose and d-xylose metabolism in Aspergillus niger. J. Gen. Microbiol. 135, 2163–2171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.