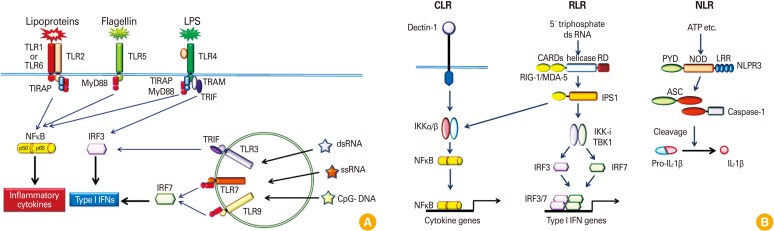
Fig. 2.
Pattern recognition receptor (PRR) and signaling (A) Toll-like receptors (TLRs). TLR receptors recognize different microbial associated molecular patterns: the heterodimer of TLR4 and MD-2 recognizes lipopolysaccharide (LPS); TLR2 recognizes triacyl and diacyl portions of lipoproteins together with TLR1 or TLR6, respectively; TLR5 recognizes flagellin; TLR3 recognizes double-stranded RNA; TLR7 recognizes single-stranded RNA and TLR9 recognizes bacterial and viral DNA, the so-called CpG DNA. The signaling pathways of TLRs are mediated by selective usage of adaptor molecules, MyD88, Toll-receptor-associated activator of interferon (TRIF), TIR-associated protein (TIRAP) and Toll-receptor-associated molecule (TRAM). (B) C-type lectins (CLRs), retinoic acid-inducible gene-like receptors (RLRs) and nucleotide binding domain (NOD)-like receptors (NLRs). CLRs recognize carbohydrates on microorganisms via the carbohydrate-binding domain. Dectin-1 is well studied. RLRs are composed of two N-terminal caspase-recruitment domains (CARDs), a central DEAD box helicase/ATPase domain, and a C-terminal regulatory domain (RD). They are localized in the cytoplasm and recognize the genomic RNA of dsRNA viruses, and dsRNA generated as the replication intermediate of ssRNA viruses. RLRs interact with IPS1 via their CARD domains, resulting in type 1 interferon production through IkB kinase, inducible (IKKi)/TANK-binding kinase 1 (TBK1). NLRs are composed of a central NOD and C-terminal leucine-rich repeats (LRRs). NODs activate caspase-1, resulting in processing of pro-interleukin-1β (IL-1β) to mature IL-1β. ASC, apoptosis-associated speck-like protein; IFN, interferon; IPS1, IFN-β promoter stimulator 1; IRF, interferon regulatory factor; MDA-5, melanoma-differentiation-associated gene 5; NF-kB, nuclear factor-kB; NLPR3, NLR family, pyrin domain-containing 3; RIG-1, retinoic acid-inducible gene I. Modified from Akira [15].