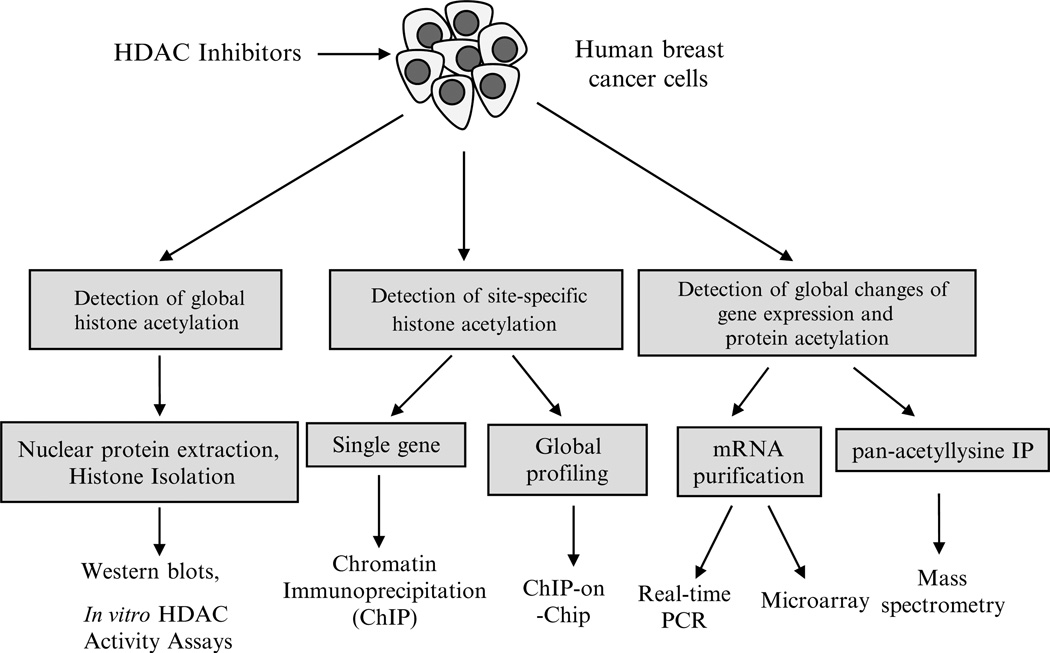
Fig. 1.
Techniques for studying HDAC inhibition in human breast cancer. After human breast cancer cells are treated with HDAC inhibitors, immunological detection methods such as Western blot or immunochemistry can be used to determine the level of histone acetylation using specific antibodies against histone H3 or H4 or specific histone lysine residues such as AcH3K9, AcH3K27, and AcH4K20. Chromatin immunoprecipitation (ChIP) is used to determine the interaction of site-specific acetylated histones with promoters of genes of interest in breast cancer cells after HDAC inhibitor treatment. DNA sequences bound to a particular acetylated histone or nonhistone protein can be isolated by ChIP and these fragments can subsequently be hybridized to a DNA microarray (such as a tiling array). This so-called ChIP-on-chip technology allows the determination of acetylated histone binding occupancy throughout the genome of the cancer cell. Quantitative-PCR is able to precisely measure the specific gene expression changes in the presence of HDAC inhibitor treatment. Microarray-based gene expression profiling can be used to identify genes whose expression is altered by HDAC inhibitor treatment. Several mass spectrometry (MS)-based proteomic methods exist to quantitatively analyze proteins that are hyperacetylated after treatment with HDAC inhibitors as well as determine isoform specific occupancy of histone modifications.