Figure 5. Phylogenetic tree of lepidopteran and other insect β-N-acetylglucosaminidases.
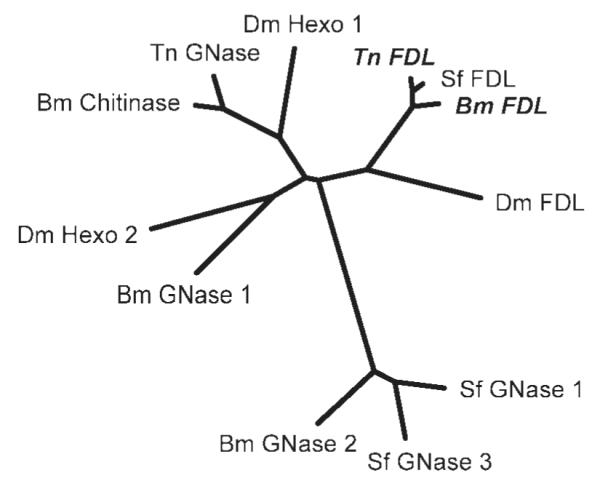
This unrooted tree demonstrates the phylogenetic relationships between cloned insect GNases. The distances between sequences are inversely proportional to their similarity. To produce this tree, a multiple sequence alignment was performed by ClustalX 2.0.10 using the default settings, which was then used to generate a distance matrix using Protdist (PHYLIP version 3.68) with the Jones-Taylor-Thornton model. An unrooted tree was then drawn by the PHYLIP drawtree postscript generator from the distance matrix using the neighbor-joining method (Neighbor, PHYLIP version 3.68). The GNase proteins and their Genbank accession numbers used in this tree are: Tn FDL (FJ695479): Trichoplusia ni processing GNase (this study), Bm FDL (FJ695481): Bombyx mori processing GNase (this study), Sf FDL (ACA30398): Spodoptera frugiperda processing GNase,21 Dm FDL (Q8WSF3): Drosophila melanogaster processing GNase,20 Bm GNase1 (BAF52531) and 2 (BAF52532): Bombyx mori non-specific GNases,27 Dm Hexo1 and 2: Drosophila melanogaster chitinolytic GNases,20 Bm Chitinase (AAC60521): Bombyx mori chitinolytic GNase,28 Tn GNase (AAL82580): uncharacterized, putative Trichoplusia ni GNase, Sf GNase 1 (ABB76924) and 3 (ABB76926): Spodoptera frugiperda non-specific GNases.22
