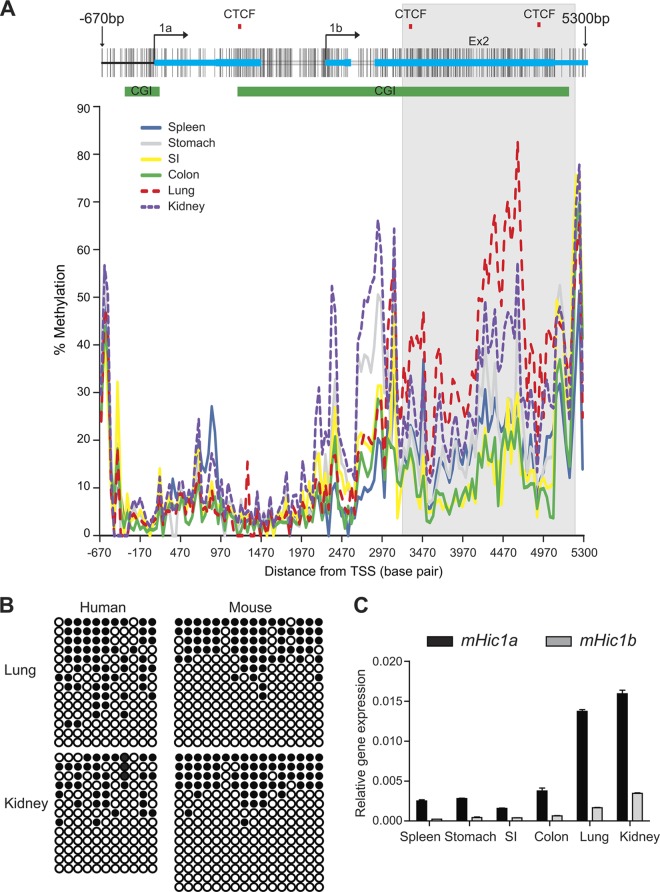
Fig 5.
Conserved role of 3′ CGI methylation in regulating gene expression. (A) The genomic structure of the mouse Hic1 gene is shown at the top. Alternative promoters at exons 1a and 1b are indicated. The green bars indicate the positions of two evolutionally conserved promoters and 3′ CGIs. The red tick marks indicate predicted CTCF-binding sites. The lower panel is a plot of methylation level versus genomic location for various mouse tissues. A 2-kb region (highlighted in gray) is differentially methylated in multiple tissues, overlaps both the 3′ CGI and two potential CTCF-binding sites, and was used for further functional characterization. Lung and kidney are hypermethylated relative to spleen, stomach, small intestine (SI), and colon tissues. (B) Comparison of tissue-specific methylation in mouse and human by clonal bisulfite sequencing indicates that Hic1 3′ CGI methylation is evolutionarily conserved. Methylation at the core regions of Hic1 3′ CGI was determined in both human (bp 4081 to 4310 relative to 1A promoter) and mouse (bp 4797 to 4947 relative to 1a promoter). Each circle represents a CpG site, and each row represents a single DNA molecule. Closed circles indicate methylation. (C) qRT-PCR shows that increased Hic1 expression from both alternate promoters correlates with 3′ CGI hypermethylation.